Abstract
Here, the complete mitogenome of Saccharina cultivar ‘Xinbenniu’ was reported. It had a circular mapping organization with a length of 37,657 bp and contained 35 protein-encoding genes, 25 transfer RNA genes (tRNAs), 3 ribosomal RNA genes (rRNAs), and 3 open reading frames (ORFs). Moreover, overall, AT content was 64.70%. The gene content and encoding order were consistent with other reported Saccharina species and cultivars. Mitogenome phylogenetic tree showed that ‘Xinbenniu’ firstly clustered together with ‘Pingbancai’ and ‘Ailunwan’ indicating their closer relationship. The complete mitogenome analysis in this work would help in understanding the genetic background of Chinese Saccharina cultivars.
Saccharina is one of the most important macroalgal genera of the order Laminariales that is economically valuable owing to its use in foodstuffs and industrial materials (Kawashima Citation2004). So far, more than 20 Saccharina cultivars have been bred and have contributed to Chinese aquaculture. ‘Xinbenniu’ was an interspecific hybrid of Saccharina japonica and Saccharina latissima which has been cultured along the coast of Shandong Province. However, its genetic information is limited. Here, we determined the complete mitogenome of ‘Xinbenniu’ (GenBank accession number MG712780) and conducted phylogenetic analysis to provide new molecular data for Saccharina cultivars
‘Xinbenniu’ samples (specimen number: 2016070051) were collected from Rongcheng, Shandong Province, China (37°15′39′′N, 122°33′56′′E) and then stored at −80 °C for the subsequent experiment. The homologous PCR amplification and primer walking strategy was used (Zhang et al. Citation2011).
The mitogenome of ‘Xinbenniu’ was a circular molecule of 37,657 bp, with a nucleotide composition of 28.41% A (10,700), 14.71%C (5,541), 20.58% G (7,750), and 36.29% T (13,666), and had an overall AT content of 64.70%. This mitogenome encoded 66 genes including three rRNAs (rnl, rns, rrn5), 25 tRNAs (trn), 35 protein-encoding genes, and 3 ORFs (orf41, orf130, and orf377). The protein-encoding region included 29,007 bp accounting for 77.03% of its mtDNA. With the exception of rpl2, rpl16, rps3, rps19, tatC, and orf130, 60 genes were encoded on H-strand. All the protein-encoding genes started with ATG codon and all of the three stop codons (TAA, TAG, TGA) were employed with a preference to TAA accounting for 68.42%. One gene cluster, composed of rps8, rpl6, rps2, rps4, was exactly conserved in ‘Xinbenniu’ and other reported Saccharina mitogenomes. Moreover, all tRNA genes were provided with standard cloverleaf secondary structures. The gene content and encoding order of ‘Xinbenniu’ mitogenome were highly conserved in the mtDNAs of these reported Saccharina species and cultivars (Yotsukura et al. Citation2010; Zhang et al. Citation2013; Zhang et al. Citation2017; Zhang et al. Citation2018).
Compared to the mitogenome of S. japonica (Accession number: AP011493), there were 13 nucleotide substitutions in ‘Xinbenniu’. Among them, eight substitutions were found in protein-encoding regions. None of the above-mentioned substitutions in protein-encoding genes caused protein variation. While comparing the mitogenomes of another two Saccharina japonica × latissima cultivars ‘Ailunwan’ (Accession number: KU556731) and ‘Pingbancai’ (Accession number: KX073817), respectively, five and four substitutions were detected in ‘Xinbenniu’ mtDNA. One intergenic region of 19 nucleotides was located between rps3 and rps9 in mitogenomes of ‘Xinbenniu’, ‘Ailunwan’, and ‘Pingbancai’ which lacked from that of ‘Rongfu’ (also a Saccharina japonica ×latissima cultivar, Accession number: JF937591).
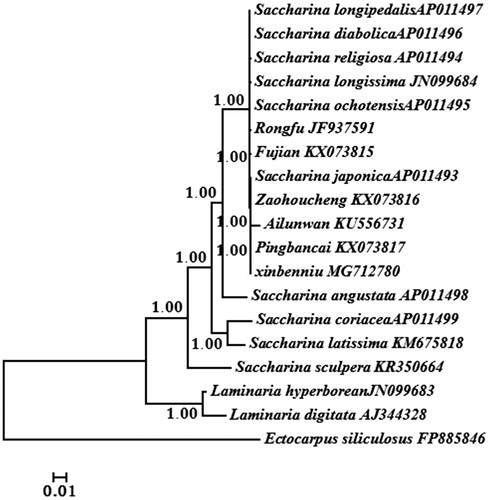
Bayesian analysis based on all protein-encoding genes of the 19 Saccharina and Laminaria algae was used to reconstruct the phylogeny via MrBayes v. 3. 1.2 (Huelsenbeck and Ronquist Citation2001). Ectocarpus siliculosus was served as the outgroup. Phylogenetic analysis showed that all algae were divided into two branches: Saccharina and Laminaria (). Six reported Chinese cultivars all belonged to the Saccharina lineage. ‘Xinbenniu’ firstly clustered with ‘Ailunwan’ and ‘Pingbancai’ supporting its genetic background. The phylogenetic tree in this work would provide more information to understand the genetic relationships of current Chinese Saccharina cultivars.
Disclosure statement
There are no conflicts of interest for all the authors including the implementation of research experiments and writing this article.
Additional information
Funding
References
- Kawashima S. 2004. Konbu. In: Ohno M, editor. Biology and technology of economic seaweeds. Tokyo: Uchida Rokakuho; p. 59–85 (in Japanese).
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics (Oxford, England). 17:754–755.
- Yotsukura N, Shimizu T, Katayama T, Druehl LD. 2010. Mitochondrial DNA sequence variation of four Saccharina species (Laminariales, Phaeophyceae) growing in Japan. J Appl Phycol. 22:243–251.
- Zhang J, Li N, Liu N, Liu T. 2017. The complete mitogenome of “Zaohoucheng”, an important Saccharina japonica cultivar in China. Mitochondr DNA B. 2:656–657.
- Zhang J, Liu N, Liu T. 2018. The complete mitogenome of “Fujian”: an important Saccharina japonica cultivar of South China. Mitochondrial DNA B. 3:588–589.
- Zhang J, Li N, Zhang ZF, Liu T. 2011. Structure analysis of the complete mitochondrial genome in cultivation variety “Rongfu”. J Ocean Univ China. 10: 351–356.
- Zhang J, Wang XM, Liu C, Jin YM, Liu T. 2013. The complete mitochondrial genomes of two brown algae (Laminariales, Phaeophyceae) and phylogenetic analysis within Laminaria. J Appl Phycol. 25:1247–1253.