Abstract
Ilex integra Thunb. is an ornamental tree distributed in Korea, Taiwan, mid-Southern China, and Japan. To understand the genetic differences between I. integra and I. cornuta, we presented complete chloroplast genome of I. integra, which is 157,548 bp long and has four subregions: 86,935 bp of large single copy (LSC) and 18,426 bp of small single copy (SSC) regions are separated by 26,093 bp of inverted repeat (IR) regions including 131 genes (86 protein-coding genes, 8 rRNAs, and 37 tRNAs). The overall GC content of the chloroplast genome is 37.6% and those in the LSC, SSC, and IR regions are 35.7%, 31.9%, and 42.9%, respectively. Phylogenetic trees show that I. integra is clustered with I. cornuta and I. latifolia. Sequence variations identified from I. integra and I. corunta presents that both species have enough sequence variations as different species.
Ilex integra Thunb., which is an ornamental tree in Ilex genus, is distributed in Korea, Taiwan, mid-Southern China, and Japan (Hu Citation1949, Citation1953). Its leaf is elliptic with entire and spineless margin, different from Ilex cornuta, giving different feeling from Ilex species used as Christmas trees. Its bark was used as birdlime to catch a bird in Japan (Divers and Kawakita Citation1888) because it secretes sticky materials on leaves and stems. In addition, several chemical analyses of I. integra has been conducted, presenting many triterpenoid saponins from leaves (Yano et al. Citation1993), triterpenes showing antifungal effects from fruits (Haraguchi et al. Citation1999), fatty acids from seeds (Hirose et al. Citation1971), and carboxyl acids from leaves and stems (Tori et al. Citation2003), which are similar to I. cornuta (Liao et al. Citation2013).
To understand differences of genetic backgrounds of I. integra and I. cornuta, total DNA of I. integra collected in Chollipo arboretum in Tae-an gun, Republic of Korea, (Voucher in InfoBoss Cyber Herbarium (IN); S. Nam, IB-00591) was extracted from fresh leaves by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq4000 at Macrogen Inc., Korea and de novo assembly and conformation were done by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation based on Ilex paraguariensis voucher Yerba mate chloroplast, complete genome (KP016928; Cascales et al. Citation2017).
The chloroplast genome of I. integra (Genbank accession is MK335537) is 157,548 bp and has four subregions: 86,935 bp of a large single copy (LSC) and 18,426 bp of small single copy (SSC) regions are separated by 26,093 bp of the inverted repeat (IR). It contained 131 genes (86 protein-coding genes, 8 rRNAs, and 37 tRNAs); 18 genes (7 protein-coding genes, 4 rRNAs, and 7 tRNAs) are duplicated in IR regions. The overall GC content of I. integra is 37.6% and those in the LSC, SSC, and IR regions are 35.7%, 31.9%, and 42.9%, respectively.
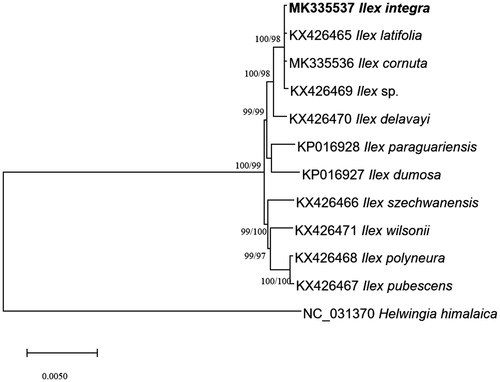
Eleven Ilex and one Helwingia chloroplast genomes from Aquifoliaceae were used for constructing neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenic trees using MEGA X (Kumar et al. Citation2018) after aligning whole chloroplast genomes using MAFFT 7.388 (Katoh and Standley Citation2013). Phylogenetic trees show that I. integra is clustered with I. cornuta from Korea, Ilex latifolia, and Ilex sp. from China (Yao et al. Citation2016) supported by high bootstrap values, agreeing with previous phylogenetic analyses based on ITS and chloroplast marker sequences (Cuénoud et al. Citation2000; Gottlieb et al. Citation2005; Jiang et al. Citation2017; ). 428 insertions and deletions and 55 single nucleotide polymorphisms identified between I. integra and I. cornuta, presents that two chloroplast genomes contain enough variations as a different species in comparison to previous studies (Duchesea chrysantha; Nymphaea alba; Coffea arabica).
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of eleven Ilex and one Helwingia chloroplast genomes from Aquifoliaceae: Ilex integra (MK335537 in this study), Ilex cornuta (MK335536), Ilex latifolia (KX426465), Ilex sp. XY-2016 (KX426469), Ilex delavayi (KX426470), Ilex paraguaiensis (KP016928), Ilex dumorsa (KP016927), Ilex szechwanensis (KX426466), Ilex wilsonii (KX426471), Ilex polyneura (KX426468), Ilex pubescens (KX426467), and Helwingia himalaica (NC_031370). Section names were displayed in the right side of phylogenetic tree (Gottlieb et al. Citation2005; Wu et al. 2008; Jiang et al. Citation2017). Phylogenetic tree was drawn based on neighbor joining tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Cascales J, Bracco M, Garberoglio MJ, Poggio L, Gottlieb AM. 2017. Integral phylogenomic approach over Ilex L. species from southern South America. Life. 7:47.
- Cuénoud P, Del Pero Martinez MA, Loizeau P-A, Spichiger R, Andrews S, Manen J-F. 2000. Molecular phylogeny and biogeography of the genus Ilex L.(Aquifoliaceae). Ann Bot. 85:111–122.
- Divers E, Kawakita M. 1888. XXIV.—On the composition of Japanese bird-lime. J Chem Soc. 53:268–277.
- Gottlieb AM, Giberti GC, Poggio L. 2005. Molecular analyses of the genus Ilex (Aquifoliaceae) in southern South America, evidence from AFLP and ITS sequence data. Am J Bot. 92:352–369.
- Haraguchi H, Kataoka S, Okamoto S, Hanafi M, Shibata K. 1999. Antimicrobial triterpenes from Ilex integra and the mechanism of antifungal action. Phytother. Res. 13:151–156.
- Hirose M, Kosuzume K, Goshima M, Tanabe E, Satoh Y, Hagitani A. 1971. The constituents of the seeds of Ilex integra Thunb. Part I. J Japan Oil Chemists' Soc. 20:7–9.
- Hu S-y. 1949. The genus Ilex in China. J Arnold Arbor. 30:233–344.
- Hu S-y. 1953. Ilex in Taiwan and the Liukiu islands. J Arnold Arbor. 34:138–162.
- Jiang L, Xu K, Fan Q, Peng H. 2017. A new species of Ilex (Aquifoliaceae) from Jiangxi Province, China, based on morphological and molecular data. Phytotaxa. 298:147–157.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Liao L, Zhou X, Liu Y-l, Xu Q-m, Li X-r, Yang S-l. 2013. Four new triterpenoidal saponins from Ilex cornuta and their cytotoxic activities. Phytochem Lett. 6:429–434.
- Tori M, Mukai Y, Nakashima K, Sono M. 2003. A carboxylic acid from Ilex integra. Molecules. 8:882–885.
- Wu Z, Raven P, Hong D. 2008. Flora of China. Vol. 11: Oxalidaceae through Aceraceae. Beijing, and Missouri: Science Press; St. Louis: Botanical Garden Press.
- Yano I, Nishiizumi C, Yoshikawa K, Arihara S. 1993. Triterpenoid saponins from Ilex integra. Phytochemistry. 32:417–420.
- Yao X, Tan Y-H, Liu Y-Y, Song Y, Yang J-B, Corlett RT. 2016. Chloroplast genome structure in Ilex (Aquifoliaceae). Sci Rep. 6:28559.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
