Abstract
The complete mitochondrial genome of Zophobas atratus (Fab.) (= Z. morio (Fab.), Z. rugipes Kirsch) is a circular molecule of 15,494 bp (GenBank accession MK140669), with 72.01% A + T content, which comprises 13 protein-coding-, 22 tRNA-, and 2 rDNA-genes. The protein-coding genes have the traditional ATN (Met) initiation codon and are terminated by the traditional stop codon, TAA, except for the nad1 and nad4 genes. A phylogenetic tree based on maximum-likelihood analysis showed that Z. atratus is closely related to Ulomoides dermestoides, an important component of traditional Chinese medicine.
The larvae of the super mealworm Zophobas atratus (Fab.) (= Z. morio (Fab.), Z. rugipes Kirsch) (Tschinkel Citation1984) is an important animal feed additive because of its protein and fat content, just like the yellow mealworm and dark mealworm (Ro and Nilsson Citation1993; Bai et al. Citation2018). There is a need, however, for more information regarding its genetic and biological background.
Samples of adult Z. atratus were obtained from our laboratory collections (maintained since March 2016) in Guiyang City, Guizhou Province, China, on 21 June 2018. Their genomic DNA was isolated and fragmented into 250 bp pieces by Covaris M220 to build a genomic library that was sequenced (pair-end 2 × 150 bp) using Illumina Hiseq X Ten (Illumina, Inc., San Diego, CA, USA). We obtained approximately 19,752 Mbp of raw data and 19,429 Mbp of clean data. The genome was assembled de novo by the SOAPdenovo v2.04 software (Luo et al. Citation2012) and the MITObim v1.8 software (https://github.com/chrishah/MITObim) (Hahn et al. Citation2013).
The Z. atratus mitochondrial (mt) genome is a 15,494 bp closed loop structure with an A/T bias and an A + T content of 72.01%. Using Perna and Kocher’s formula, the AT- and GC-skews of the major strand of the mt genome were calculated to be approximately 0.1269 and −0.3187, respectively. The length of the A/T-rich region of the mt genome was 737 bp and was located between the srRNA and tRNA-Ile.
The complete mt genome of Z. atratus contains genes coding for 13 proteins, 22 tRNAs, and 2 rRNAs, of which the 13 protein-coding genes (PCGs) had the traditional ATN (Met) start codon; of these 13 PCGs, 4 (cox1, cox2, nad5, and cob) had ATA as start codon, 6 (nad2, atp8, nad3, nad4l, nad6, and nad1) had ATT as start codon, and 3 (atp6, cox3, and nad4) had ATG as start codon. The cox1, cox2, nad2, atp6, nad5, nad4l, nad6, and cob genes had the common typical stop codon TAA; the nad1 and nad4 genes had the common stop codon TAG; and the atp8, cox3, and nad3 genes had an incomplete stop codon, T, completed by the addition of 3′ A residues to the resultant mRNA. The 22 tRNA genes identified were interspersed throughout the coding region and ranged from 58 to 70 bp in length. The size of lrRNA and srRNA genes of the Z. atratus mt genome were 1371 and 769 bp long, respectively.
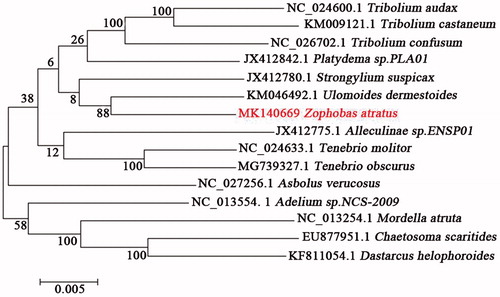
To validate the phylogenetic position of Z. atratus, 13 mitochondrial PCGs of the complete mitochondrial genomes from 15 species from Tenebrionidae were used to construct a maximum-likelihood tree using the RAxML v8.0.0 software (Stamatakis Citation2014) (). Z. atratus was closely clustered with Ulomoides dermestoides. Ulomoides dermestoides is an important component of traditional Chinese medicine (Wang et al. Citation2016), suggesting that Z. atratus may also have medicinal value besides being an important animal feed. In conclusion, the complete mtDNA of Z. atratus was decoded in this study for the first time, providing essential and important genetic and molecular data for further phylogenetic and evolutionary analysis of Tenebrionidae.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Bai Y, Li C, Yang M, Liang S. 2018. Complete mitochondrial genome of the dark mealworm Tenebrio obscurus Fabricius (Insecta: Coleoptera: Tenebrionidae). Mitochondrial DNA Part B. 3:171–172.
- Hahn C, Bachmann L, Chevreux BJNar. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. 41:e129–e129.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Ro A-I, Nilsson D-EJJoCPA. 1993. Sensitivity and dynamics of the pupil mechanism in two tenebrionid beetles. 173:455–462.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tschinkel WR. 1984. Zophobas atratus (Fab.) and Z. rugipes Kirsch (Coleoptera: Tenebrionidae) are the same species. The Coleopterists Bulletin. 38:325–333.
- Wang C-Y, Feng Y, Chen X-MJMDPA. 2016. The complete mitochondrial genome of a medicinal insect Martianus dermestoides (Coleoptera, Tenebrionidae). Mitochondr DNA A DNA Mapp Seq Anal. 27:1627–1628.