Abstract
In the present study, we determined the complete mitogenome of Opisina arenosella, the first for the family Xyloryctidae. This mitochondrial genome contains 15,389 bp, with an A + T content of 80.5% and consists of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and a control region (GenBank accession no. MK467611). The genome size, gene arrangement, A + T content, codon usage, and secondary structures of 22 transfer RNA genes of the O. arenosella mitogenome were similar to those of other sequenced Gelechioids. Bayesian analyses performed using the mitogenomes of O. arenosella and other 13 species of Gelechioidea.
The coconut black-headed caterpillar Opisina arenosella Walker is one of the major coconut leaf-feeding pests with outbreaks causing serious damage to coconut and other palms in India and Sri Lanka and it has been widely distributed in Myanmar, Pakistan, Thailand, Bangladesh, Malaysia, and Indonesia (Kumara et al. Citation2015; Shameer et al. Citation2018). Recently, it has invaded Hainan Province and Guangxi Zhuang Autonomous Region in China (Li et al. Citation2014). To date, studies of O. arenosella have mainly focused on its biological characteristics (Muralimohan and Srinivasa Citation2008; Kumara et al. Citation2015), biological control (Venkatesan et al. Citation2009; Shameer et al. Citation2018), etc. Few studies have been devoted to the molecular characterization of O. arenosella.
In this study, the complete mitochondrial genome of O. arenosella Yang is sequenced. Adult individuals were collected from Hongcheng Lake in Haikou (Hainan, China; 110°21′4′′N, 20°0′45′′E). After morphological identification, the specimens were stored at −20 °C in plant quarantine laboratory of Post-Entry Quarantine Station for Tropical Plant, Haikou Customs District, P.R. China. The hind legs of a single individual were used for DNA extraction.
We used the high-throughput sequencing method to acquire the O. arenosella complete mitochondrial genome sequences (GenBank: MK467611). The O. arenosella mitochondrial genome is 15,389 bp in length with a total A + T content of 80.5%. It contains the complete set of 37 genes which are usually found in animal mitogenomes. The gene arrangement and orientation are identical with other ditrysian Lepidoptera. The mitochondrial genome contains three tRNAs between the control region and ND2 in the order tRNA-Met, tRNA-Ile, tRNA-Glu, and for the inferred ancestral insect mitochondrial genome this order is tRNA-Ile, tRNA-Glu, tRNA-Met (Cao et al. Citation2012; Timmermans et al. Citation2014).
All PCGs start with a typical ATN codon, except COI which is started with CGA; one PCG (ND5) stop with incomplete codon TA and three PCGs (COI, COII, and ND4) stop with incomplete codon T and the remaining 9 PCGs stop with usual codon TAA. All tRNAs harbor the cloverleaf secondary structures predicted by MITOS (Bernt et al. Citation2013) except for the tRNA-Ser (AGN) lacking the DHUarm. The rrnL genes were 1364 bp in length with an A + T content of 83.5%, while the rrnS is 777 bp long with an A + T content of 85.4%. The AT-rich region is 350 bp in length, with an A + T content of 96%, with conserved motifs (‘ATAGA’ and poly-T-stretch), which are commonly observed in other lepidopteran mitochondrial genomes.
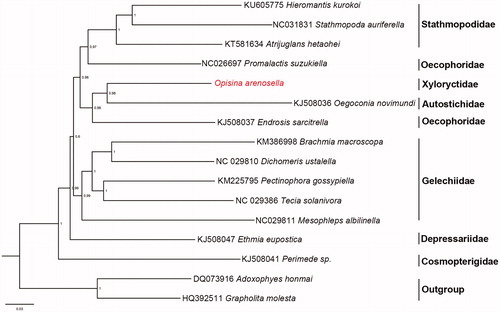
Phylogenetic analysis for O. arenosella and the other 13 Gelechioidea species was conducted based on nucleotide sequence data of 13 PCGs. Two species of Tortricoidea were used as an outgroup. The Bayesian inference (BI) method was performed through Mrbayes 3.2 (Ronquist et al. Citation2012). The resultant BI tree showed that Cosmopterigidae, Depressariidae, Gelechiidae, and Stathmopodinae form independent clades. However, Endrosis sarcitrella and Promalactis suzukiella that belong to Oecophoridae did not form monophyletic group. O. arenosella of Xyloryctidae formed a clade with E. sarcitrella of Autostichidae. These results may indicate that more mt-genome sequences are required to resolve the phylogenic relationships within Gelechioidea.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Cao YQ, Ma C, Chen JY, Yang DR. 2012. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in Lepidoptera. BMC Genomics. 13:276
- Kumara A, Chandrashekharaiah M, Subaharan K, Chakravarthy AK. 2015. Periodicity of adult emergence and sexual behaviour of coconut black headed caterpillar, Opisina arenosella Walker (Lepidoptera: Oecophoridae). Phytoparasitica. 43:701–712.
- Li HH, Yin AH, Cai B, Li WD, Lu ZS. 2014. Taxonomic status and morphology of the invasive alien species Opisina arenosella Walker (Lepidoptera: Xyloryctidae.) Chin J Appl Entomol. 51:283–291.
- Muralimohan K, Srinivasa YB. 2008. Occurrence of protandry in Opisina arenosella multivoltine moth: implications for body-size evolution. Cur Sci. 94:513–518.
- Ronquist F, Teslenko M, Mark P v d. 2012. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst Biol. 61:539–542.
- Shameer KS, Nasser M, Mohan C, Ian CW. 2018. Hardy direct and indirect influences of intercrops on the coconut defoliator Opisina arenosella. J Pest Sci. 91:259–275.
- Timmermans M, Lees DC, Simonsen TJ. 2014. Towards a mitogenomic phylogeny of Lepidoptera. Mol Phylogenet Evol. 79:169–178.
- Venkatesan T, Jalali SK, Srinivasamurthy K. 2009. Competitive interactions between Goniozus nephantidis and Bracon brevicornis, parasitoids of the coconut pest Opisina arenosella. Int J Pest Manag. 55:257–263.