Abstract
The complete mitochondrial genome of the Angolan dentex (Dentex angolensis, Poll and Maul, 1953) is 16.581 bp in length and contains 13 protein-coding genes, 22 transfer RNA genes, and 2 ribosomal RNA genes. The overall base composition of D. angolesis mtDNA is: 27.4% for A, 28% for C, 16.7% for G, 27.9% for T. Phylogenetic analysis indicated that D. angolensis is most closely related to D. tumifrons. Whole genome sequencing of D. angolensis mitochondrial DNA will contribute to improving knowledge about Sparidae evolution.
The Angolan dentex (Dentex angolensis) has one of the highest economic value among marine fishes (Decreto n. 19105, 22 Settembre 2017). For consumption purposes, it is marketed fresh, frozen, and sometimes dried salted. D. angolensis is widely distributed in the Eastern Central Atlantic along the West Coast of Africa from Morocco to Angola, including Canary Islands (Rico et al. Citation1995). The economic value of D. angolensis is driving overfishing in some regions, with substantial (10%/year) population decline (Aheto et al. Citation2012). Thus, the species is now classified as ‘Near Threatened’ in the Red List of Threatened Species (Pollard et al. Citation2014).
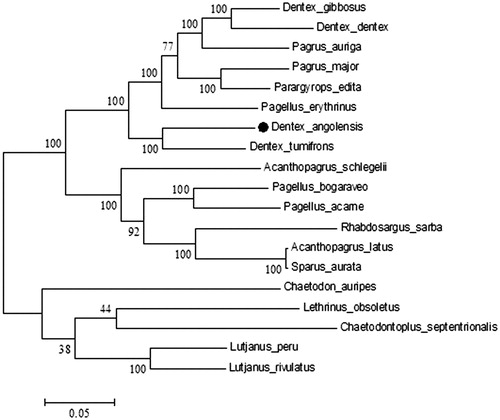
To make a step forward, our understanding of the evolution of sparid mitogenomes, we sequenced the complete mitochondrial genome of D. angolensis (GenBank accession no.MH593823). A whole specimen caught in the South Atlantic Ocean (FAO area 47; S 8°27'07.1", E 13°00'05.0") was identified as D. angolensis based on morphological features. DNA extracted was stored at the Department of Veterinary Medicine and Animal Production, University ‘Federico II’, Naples, Italy. D. angolensis mitogenome has been sequenced with Illumina HiSeq 2500 System (Illumina, San Diego, CA, USA). The complete sequence is 16.581 bp long, containing 13 protein-coding genes, 2 ribosomal RNA genes (12S rRNA and 16S rRNA), 22 transfer RNA genes (tRNA), and 2 non-coding regions (D-loop and L-origin). Mitochondrial array and gene distribution are in agreement with other vertebrate mitogenomes (Wang et al. Citation2008). Base composition is similar to one measured in other mitochondrial DNA sequences of Sparidae species, with 27.4% for A, 28% for C, 16.7% for G, 27.9% for T (Ceruso et al. Citation2018). Mitochondrial genes were distributed in two groups: most of the genes were encoded on the heavy strand, while the NADH dehydrogenase subunit 6 (ND6), and 8 tRNA genes [Gln, Ala, Asn, Cys, Tyr, Ser(UCN), Glu, Pro] are on the light strand. All protein-coding genes begin with an ATG start codon except for COI that started with GTG. Stop codons were of 4 types, i.e. TAA (ND1, ND2, ATP8, ATP6, COIII, ND4L, ND5, ND6), AGG (COI), T (COII, ND4, CYTB) and, TAG (ND3). Ribosomal subunit 12S (954 bp) was located between the tRNAPhe (GAA) and tRNALeu (TAA) genes, while 16S (1674 bp) was found between tRNAVal (TAC) and tRNALeu (TAA) genes, as in other vertebrates (Mascolo et al. Citation2018). The 22 tRNA genes vary from 66 to 74 bp in length. The control region is 904 bp long and is located between tRNAPro (TGG) and tRNAPhe (GAA). The non-coding region (L-strand origin of replication) is 35 bp long and is located between tRNAAsn (GTT) and tRNACys (GCA). To validate the phylogenetic position of D. angolensis, we constructed a phylogenetic tree using MEGA6 software (Tamura et al. Citation2013) (). The resultant phylogeny shows that D. angolensis is closely related to D. tumifrons, in agreement with Chiba et al. (Citation2009). This study can be used to evaluate genetic diversity and population structure of sparids and to provide key information for phylogenetic studies.
Figure 1. Phylogenetic analysis of D. angolensis based on the entire mtDNA genome sequences of 13 sparid fishes available in GenBank (Acanthopagrus latus NC_010977.1; Acanthopagrus schlegelii JQ_746035.1; Dentex dentex MG_727892.1; Dentex gibbosusMG_653593.1; Dentex tumifrons NC_029479.1; Pagellus acarne MG_736083.1; Pagellus bogaraveo NC_009502.1; Pagellus erythrinus MG_653592.1; Pagrus auriga AB_124801.1; Pagrus major NC_003196.1; Parargyrops edita EF_107158.1; Rhabdosargus sarba KM_272585.1; Sparus aurata LK_022698.1). Five outgroup species (Lutjanus peru KR362299.1, Lutjanus rivulatus AP006000.1, Lethrinus obsoletus NC009855, Chaetodontoplus septentrionalis NC009873 and Chaetodon auripes NC009870) were selected and the maximum likelihood method was used. Numbers above the nodes indicate 1000 bootstrap values.
Acknowledgements
The authors thank the Molecular Biology and Sequencing unit of the Stazione Zoologica Anton Dohrn for technical assistance. Celeste Mascolo has been supported by a SZN PhD fellowship.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Aheto D, Asare N, Quaynor B, Tenkorang E, Asare C, Okyere I. 2012. Profitability of small-scale fisheries in Elmina, Ghana. Sustainability. 4:2785–2794.
- Ceruso M, Mascolo C, Palma G, Anastasio A, Pepe T, Sordino P. 2018. The complete mitochondrial genome of the common dentex, Dentex dentex (Perciformes: Sparidae). Mitochondrial DNA B. 3:391–392.
- Chiba SN, Iwatsuki Y, Yoshino T, Hanzawa N. 2009. Comprehensive phylogeny of the family Sparidae (Perciformes: Teleostei) inferred from mitochondrial gene analyses. Genes Genet Syst. 84:153–170.
- Decreto n. 19105, 22 settembre 2017. Attribuzione delle denominazioni in lingua italiana delle specie ittiche di interesse commerciale. GU Serie Generale n.266 del 14-11-2017 [accessed 31 Genuary 2019].
- Mascolo C, Ceruso M, Palma G, Anastasio A, Sordino P, Pepe T. 2018. The complete mitochondrial genome of the axillary seabream, Pagellus acarne (Perciformes: Sparidae). Mitochondrial DNA B. 3:434–435.
- Pollard D, Buxton CD, Carpenter KE, Russell B. 2014. Dentex angolensis. The IUCN Red List of Threatened Species 2014: e.T170158A1283985. http://dx.doi.org/10.2305/IUCN.UK.2014-3.RLTS.T170158A1283985.en.
- Rico V, Santana JI, González JA. 1995. Ocurrence of Dentex (Polysteganus) angolensis Poll and Maul, 1953 (Sparidae) in the Canary Islands. Cybium. 19: 323–432.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang C, Chen Q, Lu G, Xu J, Yang Q, Li S. 2008. Complete mitochondrial genome of the grass carp (Ctenopharyngodon idella, Teleostei): insight into its phylogenic position within Cyprinidae. Gene. 424:96–101.
