Abstract
Sophora alopecuroides L. (Fabaceae) is an important medicinal and forage herb species with good economic value. Using an Illumina platform, we sequenced its complete chloroplast genome. Our study reveals that S. alopecuroides have a typical chloroplast genome of 154,108 bp in length, comprised of a large single-copy region (LSC) of 84,221 bp, a small single-copy region (SSC) of 18,139 bp, and inverted repeat regions (IRs) of 258,74 bp. A total of 129 genes, 81 of which are protein coding genes, 38 tRNA genes, eight rRNA genes, and two pseudogenes were identified. Phylogenetic analysis indicates that S. alopecuroides is closely related to the species of Ammopiptanthus mongolicus.
Sophora alopecuroides L. (Fabaceae) is an important medicinal and forage herb species with good economic value. It is mainly distributed in the northwest regions of China, such as Ningxia and Xinjiang. Sophora alopecuroides plays an important role in traditional Chinese medicine, owing to its anti-inflammatory and anti-tumor activities (Chang et al. Citation2014). Furthermore, it is also fine forage for increasing the weight of livestock (Fan and Wang Citation2016). However, largely due to anthropogenic cutting and climatic environment’s change, its natural habitats have been fragmented, yet none is known concerning its genetic background. In plants, chloroplast DNA (cpDNA) provided valuable phylogenetic signals for all kinds of lineages, owing to its conserved genome structures and comparatively high substitution rates (Wu and Ge Citation2012). Hence, a good knowledge of its genomic information would have made it easy to investigate genetic variations and contribute to the preservation and stabilization of local ecosystems. In the present study, we assembled and characterized the complete chloroplast genome sequence of S. alopecuroides based on the Illumina pair-end sequencing data.
The fresh leaves of a single individual of S. alopecuroides were sampled from Aletai (Xinjiang, China; 87°51′39″ E, 47°26′78″ N) and were used for the total genomic DNA extraction with the modified CTAB method (Doyle and Doyle Citation1987). DNA sample and voucher specimen of S. alopecuroides were deposited in the Molecular Ecology Laboratory (MEL), Institute of Loess Plateau, Shanxi University (Taiyuan, Shanxi, China). The whole-genome sequencing was conducted with 150 bp pair-end reads on the Illumina Hiseq Platform (Illumina, San Diego, CA). In total, about 10 million high quality clean reads were obtained and used for the cp genome assembly using Velvet software (Zerbino and Birney Citation2008). The resulting contigs were linked based on overlapping regions after being aligned to Ammopiptanthus mongolicus (KY034453) (Feng et al. Citation2017) and visualized in Geneious version 8.0.5 (Kearse et al. Citation2012). Annotation was performed with the program Plann (Huang and Cronk Citation2015) and Sequin (http://www.ncbi.nlm.nih.gov/). Together with gene annotations, the complete cp genome sequences were submitted to GenBank and deposited under the accession number MF100928. A physical map of the genome was generated by OGDRAW (http://ogdraw.mpimp-golm.mpg.de/). To confirm the phylogenetic position of S. alopecuroides, its chloroplast genome was aligned with 20 reported chloroplast genomes using MAFFT (Katoh and Standley Citation2013) and a Neighbor Joining (NJ) phylogenetic tree were constructed in MEGA6 (Tamura et al. Citation2013).
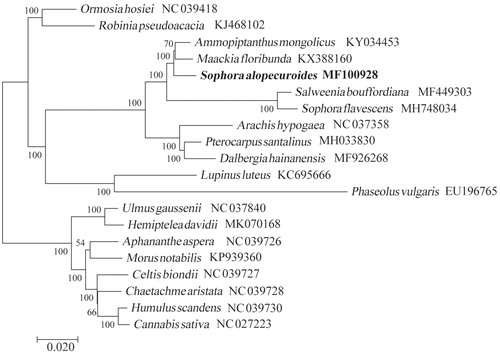
The chloroplast genome of S. alopecuroides is a typical quadripartite structure with a length of 154,108 bp, which contained inverted repeats (IR) of 25,874 bp separated by a large single-copy (LSC) and a small single copy (SSC) of 84,221 bp and 18,139 bp, respectively. The cpDNA contains 129 genes, comprising 81 protein-coding genes, 38 tRNA genes, eight rRNA genes, and two pseudogenes (rps16 and ycf1). Among the annotated genes, 10 of them contain one intron (trnK-UUU, trnV-UAC, rpoC1, atpF, trnT-CGU, trnL-UAA, petB, petD, rpl16, and ndhA), and six genes (clpP, ycf3, ndhB, rpl2, trnI-GAU, and trnA-UGC) contain two introns. The overall GC content of the plastome is 36.6% while the corresponding values of the LSC, SSC, and IR regions are 34.2%, 30.0%, and 42.8%, respectively. The phylogenetic analysis suggested that S. alopecuroides is closely related to the genera of Ammopiptanthus ().
Figure 1. The Neighbor Joining (NJ) tree based on the 20 chloroplast genomes. The bootstrap value based on 1000 replicates is shown on each node.
This complete chloroplast genome can provide valuable information to develop highly variable DNA markers for population genetic survey, species delimitation, and other ecological and evolutionary studies. This study will be fundamental to formulate potential new conservation and management strategies for this important economic value herb species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chang A, Cai Z, Wang Z, Sun S. 2014. Extraction and isolation of alkaloids of Sophora alopecuroides and their anti-tumor effects in H22 tumor-bearing mice. Afr J Tradit Complement Altern Med. 11:245–248.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fan ZC, Wang YR. 2016. The production characteristics of Sophora alopecuroides populations in different habitats. Pratacultural Sci. 33:459–470.
- Feng L, Gu LF, Luo J, Fu AS, Ding Q, Yiu SM, He JX. 2017. Complete plastid genomes of the genus Ammopiptanthus and identification of a novel 23-kb rearrangement. Conservation Genet Resour. 9:647–650.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wu ZQ, Ge S. 2012. The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Mol. Phylogenet. Evol. 62:573–578.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
