Abstract
Euphorbia hainanensis is a rare cliff top boskage, which has an important ecological and economic value. In this study, we sequenced its complete chloroplast (cp) genome using Illumina sequencing platform. The cp genome of E. hainanensis is a typical quadripartite structure with a length of 163,977 bp, which contained inverted repeats (IR) of 26,717 bp separated by a large single-copy (LSC) and a small single copy (SSC) of 92,771 bp and 17,772 bp, respectively. A total of 131 genes, 82 of which are protein coding genes, 39 tRNA genes, and eight rRNA genes were identified. The overall GC content of the plastome is 35.4% while the corresponding values of the LSC, SSC, and IR regions are 32.4%, 30.0%, and 42.4%, respectively. The phylogenetic analysis suggested that E. hainanensis was closely related to the species of E. esula.
Euphorbia hainanensis Croiz (Euphorbiaceae) is a rare cliff top boskage, which is only distributed on Changjiang County Exianling Natural Reserve in Hainan Island, China. Euphorbia hainanensis grows on the limestone peak and has an important ecological value for preventing the weathering of limestone. In addition, its roots, leaves, and bark can be used as medicines, and it has important economic value (Tian et al. Citation2015). However, because of the small number of populations and narrow habitats, E. hainanensis is treated as a rare resource species in China (Zhang et al. Citation2006). It is thus urgent to take effective measures to conserve this endemic and rare species. In plants, cpDNA provided valuable phylogenetic signals for systematic studies, owing to its conserved genome structures and comparatively high substitution rates (Wu and Ge, Citation2012). In the study, we assembled and characterized the complete chloroplast genome sequence of E. hainanensis based on the Illumina pair-end sequencing data. This will be useful for the development of conservation strategy for this rare cliff top boskage.
The fresh leaves of a wild individual of E. hainanensis were sampled from Exianling Nature Reserve (Hainan, China; 109°29′54″ E, 19°47′26″ N) and were used for the total genomic DNA extraction with a DNeasy Plant Mini Kit (QIAGEN, Valencia, California, USA). DNA sample and voucher specimen of E. hainanensis were deposited in the Molecular Ecology Laboratory (MEL), Institute of Loess Plateau, Shanxi University (Taiyuan, Shanxi, China). The whole-genome sequencing was conducted with 150 bp pair-end read on the Illumina Hiseq Platform (Illumina, San Diego, CA). In total, about 400 million high-quality clean reads is obtained and used for the cp genome de novo assembly by the program NOVOPlasty (Dierckxsens et al. Citation2017) and visualized in Geneious R8 (Kearse et al. Citation2012). Annotation was performed with the program Plann (Huang and Cronk Citation2015) and Sequin (http://www.ncbi.nlm.nih.gov/). Together with gene annotations, the complete cp genome sequences were submitted to GenBank and deposited under the accession number MH049548. And next, we generate a physical map of the genome using OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2013). A maximum likelihood (ML) tree with 1000 bootstrap replicates was inferred using RaxML version 8 (Stamatakis Citation2014) from alignments created by the MAFFT (Katoh and Standley Citation2013) using plastid genomes of 13 species.
The chloroplast genome of E. hainanensis is a typical quadripartite structure with a length of 163,977 bp, which contained inverted repeats (IR) of 26,717 bp separated by a large single-copy (LSC) and a small single copy (SSC) of 92,771 bp and 17,772 bp, respectively. The cp DNA contains 131 genes, comprising 82 protein-coding genes, 39 tRNA genes, and eight rRNA genes. Among the annotated genes, four of them contain one intron (atpF, ndhA, trnK-UUU, trnL-UAA), and six genes (ycf3, rps12, rpl2, trnI-GAU, trnA-UGC, and clpP) contain two introns. The overall GC content of the plastome is 35.4% while the corresponding values of the LSC, SSC, and IR regions are 32.4%, 30.0%, and 42.4%, respectively. The phylogenetic analysis suggested that E. hainanensis was closely related to the species of E. esula ().
Figure 1. The maximum-likelihood (ML) tree based on the 13 chloroplast genomes. The bootstrap value based on 1000 replicates is shown on each node.
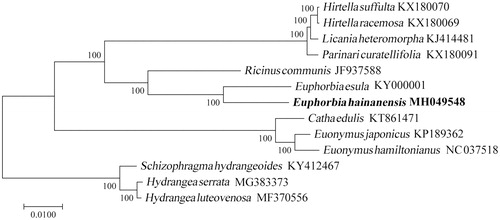
This complete cp genome can be subsequently used for population, phylogenetic, and cp genetic engineering studies of E. hainanensis, and such information would be fundamental to development potential value and formulate new conservation strategies for this rare and cliff top boskage species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:575–581.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tian XM, Li HL, He Y, et al. 2015. The GC-MS analysis for fat-soluble components for Euphorbia hainanensis latex. Northern Horticulture. 5:138–140.
- Wu ZQ, Ge S. 2012. The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Mol. Phylogenet. Evol. 62:573–578.
- Zhang RJ, QIN XS, et al. 2006. Study on the community of Euphorbia hainanensis in the limestone shrubland of Exianling Mountains, Hainan Province. Guihaia. 27:725–729.
