Abstract
The complete mitochondrial genome of a hunting spider Evarcha coreana was determined. The circular mitogenome is 14,333 bp in length (GenBank accession number MK381265), and contains 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA genes, and a putative control region. The orientation and gene order of E. coreana are identical with other spider mitogenomes. The AT content of the overall base composition is 75.86%. Twenty-two genes were in the major strand and 15 genes were in minor strand. Five intergenic regions and 25 reading frame overlaps were found in the mitogenome of E. coreana. Seven tRNA genes lost the TΨC arm stems, whereas three tRNAs lacked the dihydrouracil (DHU) arm. The control region is 697 bp in length with an A + T content of 81.06%. ATA, ATT, TTA, and TTG were initiation codons, and TAA, TAG, and T were termination codons. Phylogenetic analysis was performed using 13 PCGs with other spiders of the family Salticidae and it was seen that E. coreana is closely related to Telamonia vlijmi and Plexippus paykulli.
The hunting spider Evarcha coreana belongs to the genus of Evarcha, which includes approximately 85 species distributed across the world. These spiders predate mainly by hunting and are often found on shrubs and short plants in damp areas (Zabka Citation1985; Maddison and Hedin Citation2003). In this study, adult specimens of E. coreana were collected from Libo county, Guizhou Province, China (N25°20′, E107°53′), and were stored in the spider specimen room of Guiyang University with an accession number GYU-GZML-25.
The complete mitogenome of E. coreana (GenBank accession number MK381265) is a typical closed-circular molecule of 14,333 bp in length. It contains the entire set of 37 genes, including 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rrnL and rrnS), and a putative control region (Boore Citation1999). The overall nucleotide composition was 35.02% of A, 15.16% of G, 40.84% of T, and 8.98% of C, with a total A + T content of 75.86% that is heavily biased toward A and T nucleotides. The AT-skew and GC-skew of this mitogenome were −0.077 and 0.256, respectively. Twenty-two genes were coded on the major strand (N-strand), whereas the others were oriented on the minor strand (J-strand). The orientation and gene order of E. coreana are identical with other spider mitogenomes (Xu et al. Citation2019; Yang et al. Citation2019).
Gene overlaps in the E. coreana mitogenome were found at 25 gene junctions and involved a total of 244 bp. The longest overlap is 42 bp in length and situated between trnW and trnY. There are five intergenic spacer sequences in a total of 87 bp with length varying from 3 to 65 bp and the largest intergenic spacer is located between trnN and trnA. The length of 22 tRNAs ranged from 51 bp (trnC) to 69 bp (trnY), and nine of them were encoded on the N-strand. Ten tRNAs lack the potential to form the cloverleaf-shaped secondary structure. Seven of them (trnC, trnD, trnG, trnR, trnF, trnP, and trnL1) the TΨC arm stems, three tRNAs (trnA, trnS1 and trnS2) lost the dihydrouracil (DHU) arm. Two rRNAs have been identified on the N-strand, the rrnL gene locates between trnL1 and trnV, and the rrnS gene between trnV and trnQ. The length of rrnL and rrnS is 1009 bp and 690 bp, and their A + T content is 79.58% and 78.41%, respectively. The control region of this mitogenome is 697 bp in length with an A + T content of 81.06%, and located between the trnQ and trnM genes.
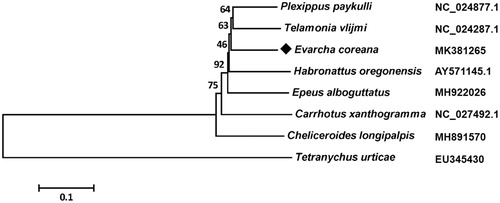
In the mitogenome of E. coreana, the total length of 13 PCGs is 10,787 bp, which accounts for 75.26% of the total genome. The A + T content of the 13 PCGs ranged from 70.87% (cox3) to 85.90% (atp8). The cox1 and cob initiated with TTA as the start codon, cox2 and cox3 began with TTG, nad2, atp6, nad4, nad4L, nad5, and nad6 started with ATA, and remaining three PCGs started with ATT. Nine PCGs including cox1, cox2, cox3, atp6, atp8, nad3, nad5, nad6, and cob are terminated with TAA as stop codon, nad1 and nad2 end with TAG, nad4 and nad4L end with a single T residue. We analyzed the amino acid sequences of 13 PCGs with neighbor-joining method to reveal the phylogenetic relationship of E. coreana with other spiders in family Salticidae. The result showed that E. coreana formed a clade with Telamonia vlijmi and Plexippus paykulli ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Boore JL. 1999. Survey and summary: animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Maddison WP, Hedin MC. 2003. Jumping spider phylogeny (Araneae: Salticidae). Invert Systematics. 17:529–549.
- Xu KK, Yang WJ, Yang DX, Li C. 2019. The complete mitochondrial genome sequence of Neoscona multiplicans (Chamberlin, 1924) (Araneae: Araneidae). Mitochondrial DNA B Resour. 4:201–202.
- Yang DX, Yan X, Xu KK, Yang WJ, Li C. 2019. The complete mitochondrial genome of Epeus alboguttatus (Araneae: Salticidae). Mitochondrial DNA B Resour. 4:316–317.
- Zabka M. 1985. Systematic and zoogeographic study on the family Salticidae (Araneae) from viet-nam. Ann Zool Warszawa. 39:197–485.