Abstract
Camellia mingii is a recently described and critically endangered new species from Southeast Yunnan Province, China. Genetic information of C. mingii would provide guidance for the conservation of this wild yellow camellia. Here, we reported and characterized its complete chloroplast (cp) genome using Illumina pair-end sequencing data. The total chloroplast genome size was 156,806 bp, including inverted repeats (IRs) of 26,020 bp, separated by a large single copy (LSC) and a small single copy (SSC) of 86,536 and 18,230 bp, respectively. A total of 132 genes, including 37 tRNA, eight rRNA, and 87 protein-coding genes were identified. Phylogenetic analysis showed that C. mingii was closely related to the clade containing C. huana and C. impressinervis.
Camellia, with ca. 120 species (Ming Citation2000; Min and Bartholomew Citation2007), mainly distribute in East and Southeast Asia. The diversity center of the genus is Southern Yangtze River of China (Ming and Zhang Citation1996). Yellow camellias are a group of species of Camellia with yellow, shiny petals, they are valuable ornamental plants and are also used in traditional Chinese medicine and commercial teas (Chang and Ren Citation1998; Min and Bartholomew Citation2007). Camellia mingii S. X. Yang is a local endemic, currently known only in Funing Xian, Yunnan Province, China. It grows on limestone in evergreen broad-leaved forests at elevation of 800–1300 m. Only three populations of C. mingii in Funing County of Yunnan were found till now, the category of Critically Endangered (CR) was recommended. Both morphological comparison and molecular analyses support C. mingii as a new member of yellow camellias (Liu et al. Citation2019). A number of chloroplast genomes of Camellia have been published (Yang et al. Citation2013; Huang et al. Citation2014); however, only few of endangered camellias were reported (Wang et al. Citation2017; Liu et al. Citation2018; Xu et al. Citation2018). In this study, we present the complete chloroplast (cp) genome sequence of C. mingii using Illumina sequencing technology.
Fresh leaves were collected from an individual of C. mingii in Funing county of Yunnan province, the voucher specimen (S. X. Yang 5610) was deposited in the Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN). Genomic DNA was isolated using a modified CTAB approach (Doyle and Doyle Citation1987). The 150 bp pair-end reads were generated using the Illumina Hi-Seq 2500 platform. Totally, 5,692,455 reads in size of 2.82 G were obtained for both ends. The chloroplast genome was de novo assembled using GetOrganelle script (Jin et al. Citation2018), with SPAdes 3.10.1 as assembler (Bankevich et al. Citation2012), then visualized by Bandage 0.8.1 (Wick et al. Citation2015) to determine paths of the cp genome. The resultant cp genome was annotated by aligning with those of its congeners (e.g. Camellia taliensis, NC022264; Camellia sinensis, KC143082) in Geneious 8.0.2 (Kearse et al. 2012). Phylogenetic analysis of 25 Camellia species and two outgroups followed our previous study (Yu et al. Citation2017).
The chloroplast genome of C. mingii was 156,806 bp, with a mean coverage of the cp genome of 80.0. The GenBank accession number of the chloroplast genome of C. mingii is MK473913. The GC content of the genome was 39.6%. The length of inverted repeats (IR), large single copy (LSC), and small single copy (SSC) was 26,020, 86,536, and 18,230 bp, respectively. The chloroplast genome of C. mingii contained 132 genes, with 8 rRNA genes, 37 tRNA genes, and 87 protein-coding genes. Annotation revealed that four rRNA genes, seven tRNA genes, and seven protein-coding genes were duplicated in the IR region.
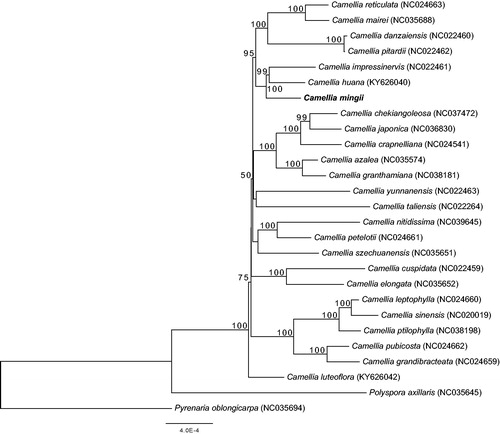
The phylogenetic relationship between C. mingii and other congeneric species was reconstructed by comparison with other 24 chloroplast genomes of Camellia. The maximum likelihood phylogenetic tree revealed that C. mingii was closely related to C. huana and C. impressinervis, both belong to Camellia subgenus Thea sect. Archecamellia (). However, the phylogenetic position of C. mingii need further investigation because of current limited sampling and the conflict between our results and the results from a previous study based on nuclear GBSSI sequences (Liu et al. Citation2019). The complete cp genome of C. mingii would be useful for the genetic diversity studies of this endangered yellow camellia species and facilitate the understanding of the conservation and restoration of C. mingii.
Figure 1. Maximum likelihood tree of the genus Camellia based on 27 complete chloroplast genome sequences, including C. mingii (GenBank ID: MK473913) sequenced in this study. The bootstrap support values are shown above the branches. Two representative taxa of Theaceae (Polyspora axillaris, NC035645; Pyrenaria oblongicarpa, NC035694) were used as outgroups.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Son P, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Chang HD, Ren SX. 1998. Theaceae (1). In: Wu CY, editor. Flora Reipublicae Popularis Sinicae. Beijing: Science Press; p. 1–251.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 19:11–15.
- Huang H, Shi C, Liu Y, Mao SY, Gao LZ. 2014. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol Biol. 14:151.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv.256479.
- Liu MM, Cao ZP, Zhang J, Zhang DW, Huo XW, Zhang G. 2018. Characterization of the complete chloroplast genome of the Camellia nitidissima, an endangered and medicinally important tree species endemic to Southwest China. Mitochondrial DNA B Resour. 3:886–887.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton Buxton, Cooper A, Markowitz S, Duran C et al. 2012. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liu ZW, Fang W, Liu ED, Zhao M, He YF, Yang SX. 2019. Camellia mingii, a new species of yellow camellias from Southeast Yunnan, China. Phytotaxa. 393:47–56.
- Min TL, Bartholomew B. 2007. Theaceae. In: Wu CY, Raven PH, editors. Flora of China. Beijing, China and Saint Louis, Missouri, USA: Science Press and Missouri Botanical Garden Press; p. 366–478.
- Ming TL. 2000. Monograph of the genus Camellia. Kunming, Yunnan: Yunnan Science and Technology Press.
- Ming TL, Zhang WJ. 1996. The evolution and distribution of genus Camellia. Acta Botanica Yunnanica. 18:1–13.
- Wang G, Luo Y, Hou N, Deng LX. 2017. The complete chloroplast genomes of three rare and endangered camellias (Camellia huana, C. liberofilamenta and C. luteoflora) endemic to Southwest China. Conservation Genet Resour. 9:583–585.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics (Oxford, England). 31:3350–3352.
- Xu XD, Zheng W, Wen J. 2018. The complete chloroplast genome of the long blooming and critically endangered Camellia azalea. Conservation Genet Resour. 10:5–7.
- Yang JB, Yang SX, Li HT, Yang J, Li DZ. 2013. Comparative chloroplast genomes of Camellia species. PLoS One. 8:e73053
- Yu XQ, Gao LM, Soltis DE, Soltis PS, Yang JB, Fang L, Yang SX, Li DZ. 2017. Insights into the historical assembly of East Asian subtropical evergreen broadleaved forests revealed by the temporal history of the tea family. New Phytol. 215:1235–1248.
