Abstract
We determined the sequence of the complete mitogenome of Bipalium kewense, an invasive land flatworm with worldwide distribution. All 37 genes encoded on this 15,666-bp genome reside on the same DNA strand. Phylogenomic analyses of mitochondrial proteins from taxa of the order Tricladida associated B. kewense with another invasive species of land planarians (Obama sp.).
The platyhelminth Bipalium kewense (family Geoplanidae) is an invasive land flatworm originating from southeastern Asia (Winsor Citation1983; Justine et al. Citation2018). Like other Bipaliinae, it features a hammerhead but is exceptional by its giant size (up to 30 cm). Remarkably, B. kewense and Bipalium adventitium were shown to produce tetrodotoxin, making these species the first known terrestrial invertebrates to possess this neurotoxin (Stokes et al. Citation2014). The current worldwide distribution of B. kewense is believed to be the result of man-mediated transportation of plants (Winsor Citation1983; Justine et al. Citation2018). Given that this invasive species preys on earthworms in cultivated areas (Justine et al. Citation2018), its dispersal has potentially devastating ecological impact. Recently, analysis of cox1 from 22 B. kewense specimens collected from Metropolitan France, other European countries, and overseas French territories on three continents revealed 100% sequence identity for this mitochondrial gene (Justine et al. Citation2018), suggesting that worldwide dispersal of a single invasive clone occurred through asexual reproduction.
We sequenced the complete mitogenome of B. kewense. DNA was obtained from a specimen that was previously sampled for cox1 analysis; this specimen collected in Ustaritz (Pyrénées-Atlantiques, France) is registered as MNHN JL184A in the collections of the Muséum National d’Histoire Naturelle (Paris, France). Sequencing was performed by the Beijing Genomic Institute (Shenzhen, China) on the BGISEQ-500 platform. A total of 60 million paired-end reads were assembled using SPAdes 3.12.0 (Bankevich et al. Citation2012) and a single mitogenome contig was recovered. Gene identification was carried out using MITOS (Bernt et al. Citation2013).
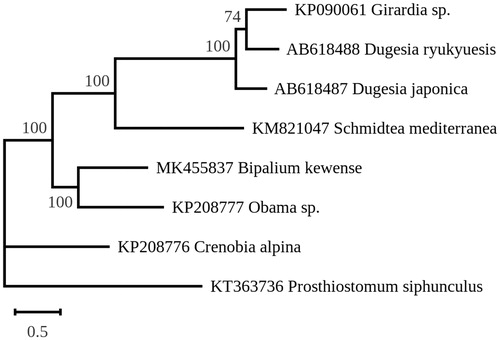
The B. kewense mitogenome (GenBank: MK455837) is 15,666 bp long and encodes 13 proteins, two rRNAs, and 22 tRNAs. All genes are located on the same DNA strand, with the protein- and rRNA- coding genes ordered as follows: cox1, ND6, ND5, cox3, atp6, ND1, cox2, ND3, ND2, rrnS, rrnL, cob, ND4L, and ND4. We inferred a maximum likelihood tree using the mitogenome-encoded proteins of B. kewense and the few Tricladida taxa whose complete mitogenomes were available (). The sequences of the proteins shared by all taxa were concatenated and analyzed with RAxML version 8.2.12 (Stamatakis Citation2014) using the MtArt model of sequence evolution (Abascal et al. Citation2007) and the Polycladida Prosthiostomum siphunculus (KT363736) as outgroup. B. kewense was found to strongly associate with the only other Geoplanidae taxon in the tree (Obama sp., most probably O. nungara), a strain from South America which is also an invasive land flatworm.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this article.
References
- Abascal F, Posada D, Zardoya R. 2007. MtArt: a new model of amino acid replacement for Arthropoda. Mol Biol Evol. 24:1–5.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Justine J-L, Winsor L, Gey D, Gros P, Thévenot J. 2018. Giant worms chez moi! Hammerhead flatworms (Platyhelminthes, Geoplanidae, Bipalium spp., Diversibipalium spp.) in metropolitan France and overseas French territories. PeerJ. 6:e4672.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Stokes AN, Ducey PK, Neuman-Lee L, Hanifin CT, French SS, Pfrender ME, Brodie ED, Brodie ED. 2014. Confirmation and distribution of tetrodotoxin for the first time in terrestrial invertebrates: two terrestrial flatworm species (Bipalium adventitium and Bipalium kewense). PLoS One. 9:e100718.
- Winsor L. 1983. A revision of the Cosmopolitan land planarian Bipalium kewense Moseley, 1878 (Turbellaria: Tricladida: Terricola). Zoo J Linn Soc. 79:61–100.