Abstract
Cordyceps tenuipes is a worldwide entomopathogenic fungus and is famous as the edible and medical value in East Asian nations. In the present study, the high-quality whole-genome of C. tenuipes was sequenced on the Illumina sequencing platform. The complete mitochondrial genome of this fungus was assembled as a single circular dsDNA of 31386 bp, including 15 protein-coding genes, 2 ribosomal RNA genes and 22 transfer RNA genes. The overall base composition of C. tenuipes is 36.6% A, 36.4% T, 11.8% C, 15.2% G, with a CG content of 27%. Phylogenetic analyses based on concatenated protein sequences from 27 taxa of five families within the order Hypocreales were conducted using Maximum likelihood (ML) and Bayesian inference (BI) methods. It is revealed that C. tenuipes is more closely related to C. militaris in the family Cordycipitaceae. This study would facilitate the future investigation of genetics, evolution and function of cordycipitoid fungi.
Cordyceps tenuipes (Peck) Kepler, B. Shrestha & Spatafora, originally classed in the genus Isaria Pers. by the name I. tenuipes Peck, is synonymous with C. takaomontana Yakush. & Kumaz. and Paecilomyces tenuipes (Peck) Samson, and has been recently combined into Cordyceps Fr. within the family Cordycipitaceae (Kepler et al. Citation2017). It is usually parasitic on lepidopteran pupae with sexual and asexual morphs, and is widely distributed throughout Asia, Europe, North America, South America, Africa, and Oceania. Studies show that C. tenuipes and its active principles possess a wide range of pharmacological actions, such as immunoregulation, anticancer, insect resistance, anti-bacteria, antivirus, anti-depression, anti-aging, antioxidant, blood sugar control, blood lipid control, and liver protection (Liang et al. Citation2018). Therefore, this fungus is commonly used as the tonic and medicine in East Asian nations (Hong et al. Citation2007). With the increasing interest in C. tenuipes both for mycology and medicine, research is necessary to get a genomic information about this potential fungus. In addition, our understanding of the genetic diversity and phylogenetic position based on mitochondrial genome (mitogenome) of C. tenuipes is lacking. This study reported the complete mitogenome of C. tenuipes assembled from Illumina sequencing data and disclosed its phylogenetic relationship to other cordycipitoid fungi.
Cordyceps tenuipes strain (YFCC 2017002) used in this study was isolated from a fruiting body of the specimen collected from Hoang Lien National Park, Sapa District, Lao Cai Province, Vietnam (22°20′N, 103°46′E, alt. 2190 m). This strain was deposited at the Yunnan Fungal Culture Collection (YFCC), the Institute of Herb Biotic Resources, Yunnan University. Mycelia cultured on PDA for 2 weeks under dark conditions were prepared to extract total genomic DNA using MiniBEST Plant Genomic DNA Extraction Kit (TaKaRa, China). Total genomic DNA was deposited at the International Joint Research Center for Sustainable Utilization of Cordyceps Bioresources in China and Southeast Asia. The whole-genome sequencing was performed on the Illumina sequencing platform (HiSeq PE150) with standard procedures. The 350 bp paired-end libraries were prepared to generate approximately 100× sequencing depth. Mitogenomic sequences of high-quality reads were assembled by SPAdes 3.9.0 with default parameter (Bankevich et al. Citation2012). The mitogenome was annotated by MFannot tool (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) and ARWEN web server, combined with manual corrections. The mitogenomic circular map of C. tenuipes was depicted by Organellar Genome DRAW tool (Lohse et al. Citation2007).
The mitogenome of C. tenuipes represents a typical hymenopteran mitogenome and is a closed loop. The annotated mitogenome of C. tenuipes was submitted to GenBank under accession no. MK234910. The total length of this circular mitogenome is 31,386 bp, containing 15 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes and 22 transfer RNA (tRNA) genes. The overall base composition is as follows: 36.6% A, 36.4% T, 11.8% C, 15.2% G, with a CG content of 27%.
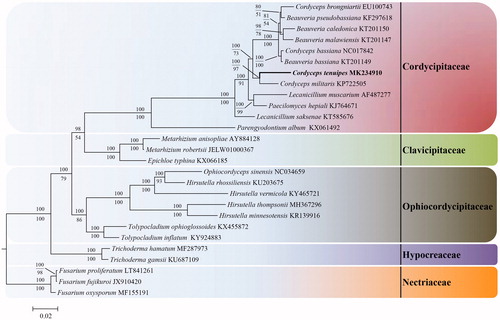
To validate the phylogenetic position of C. tenuipes, mitogenomic sequences of its 26 allied taxa were downloaded from NCBI. Mitogenomic sequences of C. tenuipes and its allies were aligned using the programme HomBlocks (Bi et al. Citation2018). Phylogenetic analyses of the concatenated protein sequences from 27 taxa were conducted using Bayesian inference (BI) and maximum-likelihood (ML) methods, employing MrBayes v.3.1.2 (Ronquist and Huelsenbeck Citation2003) and RaxML7.0.3 (Stamatakis Citation2006), respectively. The BI analysis was run on MrBayes v.3.1.2 for five million generations using the GTR + G + I model. The GTR + I model was selected as the optimal model for ML analysis, and the concatenated protein sequences were performed with 500 rapid bootstrap replicates. BI analysis is consistent with that of ML analysis from 27 taxa of the order Hypocreales. Phylogenetic tree reveals the topological structure of 27 taxa within Hypocreales and is composed of five families, namely Nectriaceae, Hypocreaceae, Ophiocordycipitaceae, Clavicipitaceae, and Cordycipitaceae (). Cordyceps tenuipes is clustered in the genus Cordyceps of Cordycipitaceae and is more closely related to C. militaris, the type species of this genus.
Figure 1. Phylogenetic relationships among 27 taxa of Hypocreales based on 14 concatenated mitochondrial protein-coding genes (PCGs). The 14 PCGs included subunits of the respiratory chain complexes (cob, cox1, cox2, and cox3), ATPase subunits (atp6, atp8, and atp9), NADH: quinone reductase subunits (nad1, nad2, nad3, nad4, nad4L, nad5, and nad6). The phylogenetic tree was generated by Bayesian inference (BI) and maximum-likelihood (ML). The Bayesian posterior probabilities and bootstraps are shown above internodes and below internodes, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SL, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Bi G, Mao Y, Xing Q, Cao M. 2018. Homblocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22.
- Hong IP, Nam SH, Sung GB, Chung IM, Hur H, Lee MW, Kim MK, Guo SX. 2007. Chemical components of Paecilomyces tenuipes (Peck) Samson. Mycobiology. 35:215–218.
- Kepler RM, Luangsa-ard JJ, Hywel-Jones NL, Quandt CA, Sung GH, Rehner SA, Aime MC, Henkel TW, Sanjuan T, Zare R, et al. 2017. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus. 8:335–353.
- Liang JD, Han YF, Tian WL, Liang ZQ, Li ZZ. 2018. Advances in bioactive components in Cordyceps takaomontana and their pharmacological effects. Acta Edulis Fungi. 25:113–119.
- Lohse M, Drechsel O, Bock R. 2007. Organellargenomedraw (ogdraw): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52:267–274.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
