Abstract
Mutations in mitochondrial DNA (mtDNA) were the important causes for non-syndromic hearing loss (NSHL). However, the molecular mechanism underlying mt-tRNA mutations and NSHL remained poorly understood. In this study, we reported here the clinical, genetic, and molecular characterization of a Han Chinese family with maternally inherited NSHL. Clinical evaluation showed a variable age at onset of NSHL. Molecular analysis of the matrilineal relatives from this family showed the presence of a novel mt-tRNAThr G15930A mutation, together with a set of genetic polymorphisms belonging to East Asia haplogroup B4b1. Interestingly, the G15930A mutation, which is localized at the highly conserved nucleotide of the anticodon stem of tRNAThr, created a novel base-pairing. Bioinformatics analysis showed that the G15930A mutation altered the secondary structure of tRNAThr and may result a failure in tRNA metabolism. As a result, the G15930A mutation may cause the mitochondrial dysfunction that was responsible for NSHL. Taken together, our data indicated that the G15930A may be a novel mutation associated with NSHL. Thus, our finding provided novel insight into the pathophysiology of maternally inherited NSHL.
Keywords:
Introduction
Mitochondrial DNA (mtDNA) mutations/variants are the important causes for non-syndromic hearing loss (NSHL) (Zheng et al. Citation2012). Of these, the 12S rRNA A1555G and C1494T mutations are found to be associated with both aminoglycoside-induced and NSHL in many families worldwide (Prezant et al. Citation1993; Fischel-Ghodsian Citation1999; Zhao et al. Citation2004). In addition to the 12S rRNA mutations, mitochondrial tRNAs (mt-tRNAs) are another hot spots for pathogenic mutations associated with deafness, for example, the common tRNASer(UCN) T7511C, tRNAGlu A14693G mutations have been identified to be associated with hearing loss (Sue et al. Citation1999; Ding et al. Citation2009).
To analysis the potential association between mt-tRNA mutations and NSHL, we initiated a mutational analysis of mt-tRNAs in NSHL patients and controls, in this study; we characterized a deafness Chinese family harbouring a novel mt-tRNAThr G15930A mutation.
Materials and methods
Subjects
We ascertained a maternally inherited Chinese family () with hearing loss from Hangzhou First people’s Hospital, Zhejiang University School of Medicine. Informed consent, blood samples, and clinical evaluations were obtained from all participating family members, under protocols approved by the Ethic Committees of Hangzhou First people’s Hospital. Moreover, 200 healthy subjects (95 females and 105 males, aged from 35 to 55 years, with an average of 45.5 years) were recruited in this study.
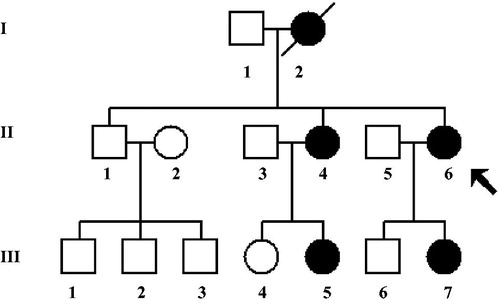
Figure 1. A three-generation Han Chinese family with hearing loss, the arrow indicates the proband, the affected members were indicated by filled symbols.
Age-appropriate audiological examinations, including pure-tone audiometry (PTA) or auditory brainstem response, immittance testing, and distortion product otoacoustic emissions, were performed. The severity of hearing impairment was classified into five grades: normal, <26 dB; mild, 26–40 dB; moderate, 41–70 dB; severe, 71–90 dB; and profound, > 90 dB.
Mutational screening of mt-tRNA genes
The Genomic DNA of each deaf patient was extracted using the Paxgene Blood DNA Isolation Kits (QIAGEN, Hilden, Germany). The 22 mt-tRNA genes were genetically amplified according to our previous investigation (Ding et al. Citation2017). The polymerase chain reaction (PCR) products were purified and analysed by the Sanger sequence, as in described elsewhere (Ding et al. Citation2018). The resultant sequence data were compared with the updated consensus Cambridge sequence (GenBank accession number: NC_012920) (Andrews et al. Citation1999).
Bioinformatics analysis
To see whether G15930A mutation altered the secondary structure of tRNAThr, we used RNA Fold Webserver to predict the thermodynamic changes of tRNAThr, according to our previous investigation (Ding and Leng Citation2012).
Genotyping analysis of GJB2 gene
The DNA fragments spanning the entire coding region of GJB2 gene were amplified by PCR using the following oligodeoxynucleotides: forward-5′-TATGACACTCCCCAGCACAG-3′ and reverse-5′-GGGCAATGCTTAAACTGGC-3′. PCR amplification and subsequent sequencing analysis were performed as detailed elsewhere (Dragomir et al. Citation2011).
Results and discussion
This Chinese family was consistent with a matrilineal inheritance, notably, four of seven matrilineal relatives suffered bilateral, late-onset, and progressive NSHL. The degree of NSHL ranged from mild to severe hearing loss, age of onset varies from 14 to 25 years. No members of this family have the history of using AmAn. The proband was a 55-year-old woman came from Hangzhou area of Zhejiang Province, she began to suffer bilateral hearing loss (90 dB right ear and 95 dB left) when she was 50 years old, moreover, we noticed that matrilineal relatives (II-4, III-5, and III-7) were also deaf patients. The PTA for II-4 was 100 dB right ear and 110 dB left ear, while the PTA for III-5 was 85 dB right ear and 100 dB left ear, the PTA for III-7 was 105 dB left ear and 75 dB left ear. The comprehensive clinical examination for each individual showed that the family exhibited the hearing loss as a sole clinical phenotype.
To see the contribution of GJB2 gene mutations to the clinical expression of hearing loss, we initiated a mutational screening for the coding region of GJB2, however, we did not detect any functional variants in GJB2 gene, suggested that nuclear gene may not play putative role in deafness expression.
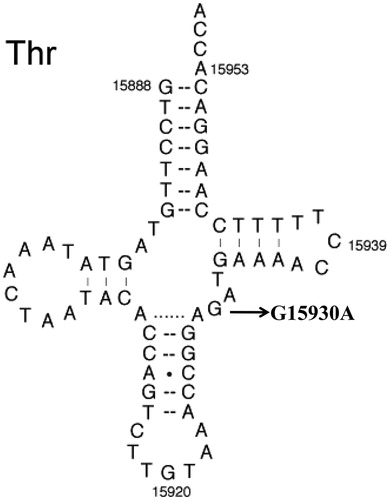
Genetic analysis suggested the occurrence of mt-tRNAThr G15930A mutation, as shown in , this mutation localized at the highly conserved nucleotide in the anticodon stem of the corresponding mt-tRNA (Salinas-Giegé et al. Citation2015), Moreover, bioinformatics analysis showed that the G15930A mutation altered the secondary structure of tRNAThr (Ding and Leng Citation2012). By molecular level, this mutation disrupted the conserved base-pairing (45G-25C) and may result the failure in mt-tRNA metabolism, this mutation only occurred in matrilineal relatives but was absent in 200 control subjects. On the other hand, the homoplasmic C3275T occurred at the same position in mt-tRNALeu(UUR) was implicated to be a pathogenic mutation associated with PCOS and metabolic syndrome (Ding et al. Citation2018). Thus, it can be speculated that G15930A mutation, which was similar to the C3275T mutation, may also cause the mitochondrial dysfunction that was responsible for deafness, therefore, we proposed that the G15930A may be a novel mutation associated with maternally inherited deafness.
Acknowledgements
The authors are grateful to the members of this family for participating for this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. 1999. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 23:147.
- Ding Y, Leng J. 2012. Is mitochondrial tRNA Leu(UUR) 3291T > C mutation pathogenic? Mitochondrial DNA. 23:323–326.
- Ding Y, Li Y, You J, Yang L, Chen B, Lu J, Guan MX. 2009. Mitochondrial tRNA(Glu) A14693G variant may modulate the phenotypic manifestation of deafness-associated 12S rRNA A1555G mutation in a Han Chinese family. J Genet Genomics. 36:241–250.
- Ding Y, Xia BH, Zhang CJ, Zhuo GC. 2017. Mutations in mitochondrial tRNA genes may be related to insulin resistance in women with polycystic ovary syndrome. Am J Transl Res. 9:2984–2996.
- Ding Y, Xia BH, Zhang CJ, Zhuo GC. 2018. Mitochondrial tRNALeu(UUR) C3275T, tRNAGln T4363C and tRNALys A8343G mutations may be associated with PCOS and metabolic syndrome. Gene. 642:299–306.
- Dragomir C, Stan A, Stefanescu DT, Savu L, Severin E. 2011. Prenatal screening for the 35delG GJB2, del (GJB6-D13S1830), and del (GJB6-D13S1854) mutations in the Romanian population. Genet Test Mol Biomarkers. 15:749–753.
- Fischel-Ghodsian N. 1999. Mitochondrial deafness mutations reviewed. Hum Mutat. 13:261–270.
- Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S, Qiu WQ, Arnos KS, Cortopassi GA, Jaber L, Rotter JI. 1993. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 4:289–294.
- Salinas-Giegé T, Giegé R, Giegé P. 2015. tRNA biology in mitochondria. Int J Mol Sci. 16:4518–4559.
- Sue CM, Tanji K, Hadjigeorgiou G, Andreu AL, Nishino I, Krishna S, Bruno C, Hirano M, Shanske S, Bonilla E, et al. 1999. Maternally inherited hearing loss in a large kindred with a novel T7511C mutation in the mitochondrial DNA tRNA(Ser(UCN)) gene. Neurology. 52:1905–1908.
- Zhao H, Li R, Wang Q, Yan Q, Deng JH, Han D, Bai Y, Young WY, Guan MX. 2004. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am J Hum Genet. 74:139–152.
- Zheng J, Ji Y, Guan MX. 2012. Mitochondrial tRNA mutations associated with deafness. Mitochondrion. 12:406–413.