Abstract
The chloroplast genome of ‘Jingxun 2,’ a new cultivar of Lavandula angustifolia, was sequenced in view of its role as an economically important aromatic plant with wide applications. De novo assembly with whole genome sequencing data completed the chloroplast genome (cp genome) of 151,459 bp in length, which included two inverted repeats (IRs) blocks of 25,467 bp, separated by the large single-copy (LSC) block of 82,945 bp and small single-copy (SSC) block of 17,580 bp. The genome encoded 123 genes consisting of 86 protein-coding genes, 29 tRNA genes, and 8 rRNA genes. The overall AT content of L. angustifolia cp genome is 62.04%. The AT contents of the LSC, SSC, and IR regions were 64.05, 68.14, and 56.66%, respectively. Further, a maximum-likelihood (ML) phylogenetic tree was reconstructed to indicate relationships among taxa Lamiaceae. The complete chloroplast genome of L. angustifolia provides utility information for aromatic plants evolutionary and genomic studies.
Lavandula angustifolia Mill., of the family Lamiaceae, is an aromatic shrub native to the Mediterranean region and cultivated worldwide both as ornamental perennials and crops (Upson and Andrews Citation2004). These plants have been used either dried or to extract essential oil for centuries for many therapeutic and cosmetic purposes. Lavandula angustifolia was introduced into China in the 1950s and becomes a popular crop with expanding cultivated area to over 3000 ha in Xinjiang Province. ‘Jingxun 2’ is a new lavender cultivar which was bred recently (Bai et al. Citation2015). With its high linalool/linalyl acetate and low camphor, the essential oils of this cultivar are among the finest and most desired lavender oils in China.
Samples of ‘Jingxun 2’ were collected from Xinjiang, Yili (43°50′9.66″N, 81°10′21.73″E) and kept at the Chinese national herbarium, Institute of Botany, Chinese Academy of Sciences (voucher specimen: 02308796). Total DNA was extracted via modified cetyltrimethylammonium bromide (CTAB) method (Doyle Citation1987) and then sequenced by MiSeq following the manufacturer’s instructions. Approximately 1019 Mb of clean data were obtained and de novo assembled by SOAPdenovo version 2.04 (Luo et al. Citation2012). All genes were further confirmed and manually adjusted by Basic Local Alignment Search Tool (BLAST) searches. The genome of 151,459 bp in length is featured with the conserved quadripartite structure of chloroplast containing a large single copy region (LSC) with a size of 82,945 bp, a small single copy region (SSC) with a size of 17,580 bp and two inverted repeat regions (IRA and IRB) each with the size of 25,467 bp. Annotation of the plastome showed that it contains 123 genes, of which 86 were protein-coding genes (79 PCG species), 29 were transfer RNA genes (23 tRNA species), and 8 were ribosomal RNA genes (4 rRNA species). Among these genes, 16 genes were duplicated in the two IR regions, including 7 PCG species (rpl2, rpl23, ycf2, ycf15, ndhB, rps7, and rps12), 5 tRNA species (trnI-CAT, trnL-CAA, trnV-GAC, trnR-ACG, and trnN-GTT), and 4 rRNA species (rrn4.5, rrn5, rrn16, and rrn23). Five genes (rpoC1, ndhK, rpl2, ndhB, and rps12) harboured a single intron whereas only ycf3 had two introns. The overall AT content of L. angustifolia cp genome is 62.04%, while the corresponding AT contents of the LSC, SSC, and IR regions are 64.05, 68.14, and 56.66%, respectively.
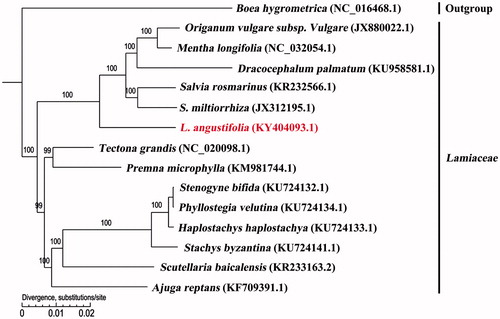
To validate the phylogenetic position of L. angustifolia, a maximum-likelihood (ML) phylogenetic tree was reconstructed with MEGA version 6 (Tamura et al. Citation2013) using the 14 complete chloroplast genomes of Lamiaceae (). The phylogenetic analysis showed that all representatives of Lamiaceae were clustered into two monophyletic clades with high bootstrap support values. Origanum vulgare, Mentha longifolia, Dracocephalum palmatum, Rosmarinus officinalis, and Salvia miltiorrhiza were clustered together firstly both belongs to Mentheae, then clustered with L. angustifolia, which belongs to Ocimeae. Overall, the complete chloroplast genome reported here provided a useful tool not only for population genetics works of L. angustifolia, but also for phylogenetic and evolutionary studies within Lamiaceae.
Figure 1. Molecular phylogeny of L. angustifolia and other 13 related species in Lamiaceae rooted by Boea hygrometrica based on plastome sequence data. The complete chloroplast genome is downloaded from NCBI database and the phylogenic tree is constructed by MEGA6 software. The gene’s accession number for tree construction is listed as follows: Boea hygrometrica (NC_016468.1), Origanum vulgare subsp. Vulgare (JX880022.1), Mentha longifolia (NC_032054.1), Dracocephalum palmatum (KU958581.1), Salvia rosmarinus (KR232566.1), S. miltiorrhiza (JX312195.1), L. angustifolia (KY404093.1), Tectona grandis (NC_020098.1), Premna microphylla (KM981744.1), Stenogyne bifida (KU724132.1), Phyllostegia velutina (KU724134.1), Haplostachys haplostachya (KU724133.1), Stachys byzantina (KU724141.1), Scutellaria baicalensis (KR233163.2), and Ajuga reptans (KF709391.1). Lavandula angustifolia is labelled with red bold in the study. Bootstrap values are shown as percentages.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bai HT, Shi L, Li H, Liu JQ, Han H. 2015. A new lavender cultivar ‘Jingxun 2’. Acta Hortic Sin. 42:2971–2972.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 1:18.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Upson T, Andrews S. 2004. The genus Lavandula. Portland (OR): Timber Press.
