Abstract
The complete mitochondrial genome (mitogenome) of Ostrinia palustralis memnialis Walker, 1859 (Lepidoptera: Crambidae) was determined to be 15,246 bp with a typical set of genes (13 protein-coding genes [PCGs], 2 rRNA genes, and 22 tRNA genes) and one non-coding region, with an arrangement identical to that observed in most lepidopteran genomes. Twelve PCGs have the typical ATN start codon, whereas COI has an atypical CGA codon frequently found in the start region of the lepidopteran COI. At 330 bp, the A + T-rich region is well within the range found in other Pyraloidea. The A + T content of the whole genome, PCGs, srRNA, lrRNA, tRNAs, and the A + T-rich region all are well within the range found in other Pyraloidea. Phylogenetic analyses of the concatenated sequences of the 13 PCGs using Bayesian inference (BI) and maximum-likelihood (ML) methods placed O. palustralis as a sister group to Ostrinia furnacalis and Ostrinia nubilalis, with the highest nodal support. The subfamilies within Crambidae, such as Nymphulinae, Spilomelinae, and Pyraustinae, all formed monophyletic groups with the highest nodal support.
Ostrinia palustralis memnialis is an oligophagous insect that feeds on dock plants, Rumex spp. (Polygonaceae) (Mutuura and Munroe Citation1970; Kwon et al. Citation2006). The eggs of this subspecies are pale yellow, and the larvae are usually reddish black (Kwon et al. Citation2006). The pupae are reddish dark brown and pupate inside stems and roots (Kwon et al. Citation2006). The species pupates once a year, overwintering as larvae or pupae inside roots, and adults emerge during the first week of June to late August in Korea (Kwon et al. Citation2006). The species has a transpalaearctic distribution, and is found from Sweden south to Italy east through Eastern Europe to Russia, and in all regions of Asia (Shin Citation2001).
An adult male O. palustralis memnialis was collected from Guleopdo Island, near Incheon Metropolitan City, South Korea (37°11′28.7′′N, 125°58′47.9′′E) in 2016. This voucher specimen was deposited at the Chonnam National University, Gwangju, Korea, under the accession number CNU6788. Using DNA extracted from the hind legs, three long overlapping fragments (LFs; COI-ND4, ND5-lrRNA, and lrRNA-COI) were amplified using three sets of primers that had been published previously (Kim et al. Citation2012). Subsequently, these LFs were used as templates to amplify 26 short fragments (Kim et al. Citation2012).
A phylogenetic tree was constructed with Bayesian inference (BI) and maximum-likelihood (ML) methods using the CIPRES Portal version 3.1 (Miller et al. Citation2010). This analysis was based solely on the concatenated nucleotide sequences of 13 PCGs, because srRNA sequence information was not available for other species of Ostrinia. An optimal substitution model (GTR + Gamma + I) was determined by a comparison of AIC scores (Akaike Citation1974) using Modeltest version 3.7 (Posada and Crandall Citation1998).
The complete 15,246 bp mitogenome of O. palustralis memnialis is composed of typical sets of genes (two rRNAs, 22 tRNAs, and 13 PCGs) and a major non-coding 330 bp A + T-rich region (GenBank accession no. MH574940). The gene arrangement of O. palustralis memnialis is identical to that of the Ditrysia (Lepidoptera), which have the order trnM-trnI-trnQ (where the underlining indicates a gene inversion) between the A + T-rich region and ND2 (Kim et al. Citation2010), instead of the ancestral trnI-trnQ-trnM order found in the majority of insects (Boore Citation1999). Twelve PCGs had the typical ATN start codon, whereas COI had an atypical CGA codon frequently found in the start region of the lepidopteran COI (Kim et al. Citation2012).
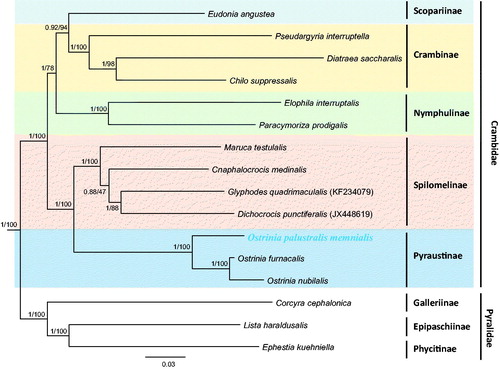
A phylogenetic analysis with concatenated sequences of the 13 PCGs revealed a sister relationship between O. palustralis memnialis and the sister group, Ostrinia Furnacalis, and Ostrinia nubilalis, all of which were confirmed to belong to Ostrinia with the highest nodal support (Bayesian posterior probabilities [BPP] = 1; Bootstrap support [BS] = 100) (). This result is consistent with that of a previous phylogenetic study using morphology and COII sequences, in that O. palustralis was confirmed as belonging to a different group than O. furnacalis and O. nubilalis (Ishikawa et al. Citation1999). The subfamilies within Crambidae, such as Nymphulinae, Spilomelinae, and Pyraustinae, all formed monophyletic groups with the highest nodal support (BPP = 1; BS = 100). Currently, about 14 subfamilies are recognized in Crambidae (Nuss et al. Citation2003–2017), but only five subfamilies have mitogenome sequences available. Therefore, additional mitogenome sequences from diverse taxonomic groups are required to elucidate the relationships of the subfamilies in Crambidae.
Figure 1. Bayesian inference (BI) method-based phylogenetic tree constructed for Crambidae in Lepidoptera, including Ostrinia palustralis memnialis, using the concatenated sequences of 13 PCGs. The numbers at each node indicate the Bayesian posterior probabilities (first) using the BI method and the bootstrap support (second) using the ML method. The scale bar indicates the number of substitutions per site. Corcyra cephalonica, Lista haraldusalis, and Ephestia kuehniella from the family Pyralidae were used as the outgroups. GenBank accession numbers are as follows: Eudonia angustea, KJ508052 (Timmermans et al. Citation2014); Pseudargyria interruptella, KP071469 (Unpublished); Diatraea saccharalis, FJ240227 (Li et al. Citation2011); Chilo suppressalis, HQ860290 (Yin et al. Citation2011); Elophila interruptalis, KC894961 (Park et al. Citation2014): Paracymoriza prodigalis, JX144892 (Ye et al. Citation2013); Maruca testulalis, KJ623250 (Zou et al. Citation2016); Cnaphalocrocis medinalis, JQ647917 (Wan et al. Citation2013); Glyphodes quadrimaculalis, KF234079 (Park et al. Citation2015); Dichocrocis punctiferalis, JX448619 (Wu et al. Citation2013); O. p. memnialis, MH574940 (this study); O. furnacalis, AF467260 (Coates et al. Citation2005); O. nubilalis, AF442957 (Coates et al. Citation2004); Corcyra cephalonica, HQ897685 (Wu et al. Citation2012); Lista haraldusalis, KF709449 (Ye et al. Citation2015); and Ephestia kuehniella, KF305832 (Traut et al. Citation2013).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Akaike H. 1974. A new look at the statistical model identification. IEEE Trans Autom Control. 19:716–723.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Coates BS, Sumerford DV, Hellmich RL. 2004. Geographic and voltinism differentiation among North American Ostrinia nubilalis (European corn borer) mitochondrial cytochrome c oxidase haplotypes. J Insect Sci. 308:258–260.
- Coates BS, Sumerford DV, Hellmich RL, Lewis LC. 2005. Partial mitochondrial genome sequences of Ostrinia nubilalis and Ostrinia furnicalis. Int J Biol Sci. 1:13–18.
- Ishikawa Y, Takanashi T, Kim CG, Hoshizaki S, Tatsuki S, Huang Y. 1999. Ostrinia spp. in Japan: their host plants and sex pheromones. Entomol Exp Appl. 91:237–244.
- Kim JS, Park JS, Kim MJ, Kang PD, Kim SG, Jin BR, Han YS, Kim I. 2012. Complete nucleotide sequence and organization of the mitochondrial genome of eri-silkworm, Samia cynthia ricini (Lepidoptera: Saturniidae). J Asia Pac Entomol. 15:162–173.
- Kim MJ, Wan X, Kim KG, Hwang JS, Kim I. 2010. Complete nucleotide sequence and organization of the mitogenome of endangered Eumenis autonoe (Lepidoptera: Nymphalidae). Afr J Biotechnol. 9:735–754.
- Kwon O, Park J, Lee IY, Park JE. 2006. General biology of Ostrinia palustralis memnialis (Walker), a potential biological control agent of Rumex spp. in Korea. Entomol Res. 36:179–182.
- Li WW, Zhang XY, Fan ZX, Yue BS, Huang FN, King E, Ran JH. 2011. Structural characteristics and phylogenetic analysis of the mitochondrial genome of the sugarcane borer, Diatraea saccharalis (Lepidoptera: Crambidae). DNA Cell Biol. 30:3–8.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the 9th Gateway Computing Environments Workshop (GCE). New Orleans (LA): IEEE; p. 1–8.
- Mutuura A, Munroe E. 1970. Taxonomy and distribution of the European corn borer and allied species: genus Ostrinia (Lepidopiera: Pyraddae). Mem Entomol Soc Can. 102:1–112.
- Nuss M, Landry B, Mally R, Vegliante F, Tränkner A, Bauer F, Hayden J, Segerer A, Schouten R, Li H, et al. 2003–2017. Global information system on pyraloidea. [accessed 2018 Aug 7]. http://www.pyraloidea.org/
- Park JS, Kim MJ, Ahn SJ, Kim I. 2015. Complete mitochondrial genome of the grass moth Glyphodes quadrimaculalis (Lepidoptera: Crambidae). Mitochondrial DNA. 26:247–249.
- Park JS, Kim MJ, Kim SS, Kim I. 2014. Complete mitochondrial genome of an aquatic moth, Elophila interruptalis (Lepidoptera: Crambidae). Mitochondrial DNA. 25:275–277.
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics. 14:817–818.
- Shin YH. 2001. Coloured illustrations of the moths of Korea. Seoul, South Korea: Academy Book Publishing; p. 160.
- Timmermans MJ, Lees DC, Simonsen TJ. 2014. Towards a mitogenomic phylogeny of Lepidoptera. Mol Phylogenet Evol. 79:169–178.
- Traut W, Vogel H, Glöckner G, Hartmann E, Heckel DG. 2013. High-throughput sequencing of a single chromosome: a moth W chromosome. Chromosome Res. 21:491–505.
- Wan X, Kim MJ, Kim I. 2013. Description of new mitochondrial genomes (Spodoptera litura, Noctuoidea and Cnaphalocrocis medinalis, Pyraloidea) and phylogenetic reconstruction of Lepidoptera with the comment on optimization schemes. Mol Biol Rep. 40:6333–6349.
- Wu QL, Gong YJ, Shi BC, Gu Y, Wei SJ. 2013. The complete mitochondrial genome of the yellow peach moth Dichocrocis punctiferalis (Lepidoptera: Pyralidae). Mitochondrial DNA. 24:105–107.
- Wu YP, Li J, Zhao JL, Su TJ, Luo AR, Fan RJ, Chen MC, Wu CS, Zhu CD. 2012. The complete mitochondrial genome of the rice moth, Corcyra cephalonica. J Insect Sci. 12:72.
- Ye F, Shi Y, Xing L, Yu H, You P. 2013. The complete mitochondrial genome of Paracymoriza prodigalis (Leech, 1889) (Lepidoptera), with a preliminary phylogenetic analysis of Pyraloidea. Aquat Insect. 35:71–88.
- Ye F, Yu HL, Li PF, You P. 2015. The complete mitochondrial genome of Lista haraldusalis (Lepidoptera: Pyralidae). Mitochondrial DNA. 26:853–854.
- Yin J, Wang AM, Hong GY, Cao YZ, Wei ZJ. 2011. Complete mitochondrial genome of Chilo suppressalis (Walker) (Lepidoptera: Crambidae). Mitochondrial DNA. 22:41–43.
- Zou Y, Ma W, Zhang L, He S, Zhang X, Zeng T. 2016. The complete mitochondrial genome of the bean pod borer, Maruca testulalis (Lepidoptera: Crambidae: Spilomelinae). Mitochondrial DNA A. 27:740–741.
