Abstract
The mitochondrial genome sequence of Minous monodactylus (16,459 bp) was determined by the MiSeq platform for the first time in the genus Minous. It encoded the 37 genes (13 protein-coding genes, 22 tRNAs, and two ribosomal RNAs) and two conserved noncoding regions; control region (D-loop) and the origin of light strand (OL). Twenty-eight genes were encoded on the heavy (H) strand, while eight tRNAs and ND6 genes were on the light (L) strand. According to the phylogenetic analysis with other complete mitogenomes from order Scorpaeniformes, M. monodactylus was clustered with Synanceia verrucosa as family Synanceiidae with 81% sequence identity.
The grey stingfish, Minous monodactylus (Scorpaeniformes: Synanceiidae) is a venomous demersal marine fish species, which inhabits the bottoms of the continental shelf (Yamada et al. Citation1995). It is widely distributed in the Indo-West Pacific region including East Africa, Red Sea, Indonesia, and Japan (Eschmeyer and Hallacher Citation1979). Although 12 species are currently reported in genus Minous based on the FishBase (http://www.fishbase.org), no complete mitochondrial genome in the genus is documented. We here report the mitochondrial genome of M. monodactylus, which was collected from Korean water as the first mitogenome sequence in the genus Minous.
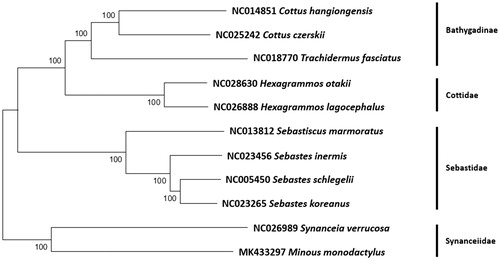
The specimen of M. monodactylus was collected during a regular EEZ fish survey (32°03′28.3′′N, 125°05′58.6′′E) by the National Institute of Fisheries Science (NIFS) in 2013. The specimen was confirmed by both the morphological characteristics and its COI sequence identity with GenBank database (KP330538). The specimen and its DNA were stored at Pukyong National University, Korea and its mitochondrial genome were determined by next-generation sequencing (NGS) technique. The mitochondrial DNA extracted by the commercial purification kit (Abcam, Cambridge, UK) was fragmented into smaller sizes (∼350 bp) by Covaris M220 Focused-Ultrasonicator (Covaris Inc., Woburn, MA). A library was constructed by TruSeq® RNA library preparation kit version 2 (Illumina Inc., San Diego, CA), which was further sequenced by MiSeq sequencer (Illumina Inc., San Diego, CA). Geneious® version 11.0.2 (Kearse et al. Citation2012) was used for mitogenome assembly, and phylogenetic analysis was conducted by MEGA version 7.0 program with minimum evolutionary (ME) algorithm (Kumar et al. Citation2016) ().
Figure 1. Phylogenetic tree of Minous monodactylus within the Scorpaeniformes. Phylogenetic tree of M. monodactylus complete genome was constructed by MEGA7 software with minimum evolution (ME) algorithm with 1000 bootstrap replications. GenBank accession numbers were shown followed by each species scientific name.
The circular mitochondrial genome of M. monodactylus (MK433297) was 16,459 bp in length, which encodes the canonical 37 genes (13 protein-coding genes, 22 tRNAs, and two ribosomal RNAs). The A + T contents were 55.5%, which was higher than G + C composition (44.5%). Twenty-eight genes were located on the heavy (H) strand, while nine were located on the light (L) strand. The control region (D-Loop) was located between tRNA-Pro and tRNA-Phe while origin of light strand (OL) was located between tRNA-Asn and tRNA-Cys. Except for COX1 (GTG), ATG was predicted as the start codon in all the protein-coding genes. The incomplete stop codon (T−/TA−) was identified in six genes including ND2, COX2, COX3, ND3, ND4, and Cyt b. Three tRNA clusters (IQM, WANCY, and HSL) were conserved. Except for in tRNA-Ser(GTC), which lacks the dihydrouridine (DHU) arm, the typical cloverleaf secondary structures were predicted in all the tRNAs according to the result by ARWE software (Canbäck and Laslett Citation2008).
The mitogenome of M. monodactylus showed the highest sequence identity (81%) to Synanceia verrucosa (KP789313), the only synanceiid mitogenome sequence in the GenBank database. The phylogenetic analysis with mitogenomes in Scorpaeniformes also showed that two synanceiid species, S. verrucosa, and M. monodactylus were clustered together distinct from those in the others. Additional mitogenome sequences should be supplemented to have better knowledge about the evolutionary relationship in the family Synanceiidae.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Canbäck B, Laslett D. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Eschmeyer WN, Hallacher LE. 1979. The scorpionfish genus Minous (Scorpaenidae, Minoinae) including a new species from the Indian Ocean. Proceedings of the California Academy of Sciences/California Academy of Sciences; Material sin tematizar Bibliografía: p.470–473.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Yamada U, Shirai S, Irie T. 1995. Names and illustrations of fishes from the East China Sea and the yellow sea: Japanese Chinese Korean. Tokyo, Japan: Overseas Fishery Co-operation Foundation.
