Abstract
Morphological characters and distribution pattern of the snake eel, Bascanichthys deraniyagalai is debatable during the past half-century. Thus, the species is re-described herewith by morphometry, vertebrae, and molecular data. In recent Chilika expedition in India, we have collected 10 specimens and observed a vertebral count dimorphism. The present study distinctly detected two vertebral groups of B. deraniyagalai: first group with 5 predorsal vertebrae, 81–83 preanal vertebrae, 190–196 total vertebrae in females; however, the sex of the second group with 5 predorsal vertebrae, 72–74 preanal vertebrae, 176–178 total vertebrae was not able to confirm. Further, the molecular data of morphologically identified two distinct groups of B. deraniyagalai shows negligible Kimura 2 parameter genetic divergence and cohesive clustering in Neighbor-Joining phylogeny proved to be the two different morphs of the same species. Hence, the present study indicated there might be vertebral count or sexual dimorphism in B. deraniyagalai, which need further sampling and taxonomic revision to unwrap the fact.
Introduction
Snake eels or worm eels of family Ophichthidae are distributed worldwide from tropical to warm-temperate oceans, and they live in shallow water to deep water at a depth of 1300 m (McCosker Citation2010). The family Ophichthidae is represented by a total of 344 valid species worldwide containing two subfamilies Myrophinae (69 species) and Ophichthinae (275 species) worldwide (Eschmeyer and Fong Citation2018). The snake eel, Bascanichthys deraniyagalai Menon, is described under subfamily Ophichthinae, family Ophichthidae, and order Anguilliformes. The species was described with five specimens collected from Arasalar River in Tamil Nadu state of Southern India. However, during the original description of this species, the characteristics of vertebral pattern and key characters are being overlooked.
In recent past, the molecular data have gained evidence as supporting tool in morphology as well as in systematics research; that enables unequivocal species identification (Laskar et al. Citation2018). The short fragments of mitochondrial as well as nuclear gene have been tested to estimate the phylogenetic relationship of eel species and revised taxonomic hierarchy (Jamandre et al. Citation2007; Tang and Fielitz Citation2012; Peninal et al. Citation2017). The molecular study also revealed the timescale for origin as well as diversification of eel species and evidenced the major lineages originated between the end of the Cretaceous and Early Eocene (Santini et al. Citation2013). Further, the partial segment of mitochondrial Cytochrome C oxidase subunit I gene has been proved to be effective in species identification of several other faunal groups including both freshwater and marine fishes (Mohapatra et al. Citation2013; Laskar et al. Citation2018). Despite the fact, many new Anguilliformes fishes have been recently discovered form India (Ray et al. Citation2015; Mohapatra et al. Citation2016, Citation2017a, Citation2017b, Citation2017c), but very few genetic information are available in both GenBank and BOLD database. The present study was shouted to re-examine the poorly described B. deraniyagalai by using morphology, meristic, vertebrae, and molecular data to re-describe the species.
Materials and methods
Sample collection and morphological data
Total 10 specimens were collected using box trap net (locally known as Khanda) operated by the local fishermen in Chilika lagoon at the outside of the Nalban Bird Sanctuary (19.69 N 85.29 E). The specimens were collected from the scattered sea grasses in the box trap net, and immediately preserved in 70% ethanol. The specimens were deposited in Estuarine Biology Regional centre, Zoological Survey of India, Gopalpur-on-Sea (Registration No. EBRC/ZSI/F9312). Further, the specimens were compared with the original description as well as the holotype and paratypes (ZSI F1964/2, ZSI F1965/2) and confirmed as B. deraniyagalai. Vertebrae were counted from radiographs as explained by Bohlke (Citation1982). The holotype of the species was also re-examined for the vertebral count, morphometric data, and meristic characters and thus, evaluated and compared. The tissue samples were collected into the 500 µl commercialized ATL buffer (Qiagen, Valencia, CA). The samples were stored at –20 °C in Centre for DNA Taxonomy laboratory, ZSI, Kolkata for further molecular analysis.
DNA isolation, PCR and sequencing
The total genomic DNA was extracted followed by QIAamp DNA Mini Kit (Qiagen, Valencia, CA) standard protocol. The published primer pair, FishF1-5′TCAACCAACCACAAAGACATTGGCAC3′, and FishR1-5′TAGACTTCTGGGTGGCCAAAGAATCA3′ (Ward et al. Citation2005) was used for amplification of partial mitochondrial cytochrome c oxidase subunit I (mtCOI) (∼650 bp) gene segment in a Veriti® Thermal Cycler (Applied Biosystems, Foster City, CA). The 25 µl PCR mixture contains 10 pmol of each primer, 100 ng of DNA template, 1× PCR buffer, 1.0–1.5 mM of MgCl2, 0.25 mM of each dNTPs, and 0.25 U of Platinum Taq DNA Polymerase High fidelity (Invitrogen, Life Science Technologies). PCR conditions were initial denaturation at 94 °C (2 min) followed by 30 cycles at 94 °C (45 s), 50 °C (45 s), and 72 °C (1 min), and a final elongation at 72 °C (8 min). The PCR amplified products were checked in 1% agarose gel containing ethidium bromide (10 mg/ml). Further, the PCR products were purified using QIAquickR Gel extraction kit (QIAGEN Inc., Germantown MD), and cycle sequencing products were cleaned by using standard BigDye X Terminator Purification Kit (Applied Biosystems, Foster City, CA). Sequencing was done bi-directionally in 48 capillary array 3730 DNA Analyzer (Applied Biosystems, Foster City, CA) following Sanger sequencing methods in the in-house sequencing facilities in ZSI, Kolkata.
Sequence quality control measures, dataset preparation and analysis
The generated chromatograms that represent sequences of both DNA strands were obtained for each sample. The noisy sequences were trimmed at both end and >2% ambiguous bases were discarded from the generated chromatograms, using the quality value of >40 for bidirectional reads. The sequences were submitted to GenBank database for acquiring the accession numbers. The homology search of the generated sequences was performed through nucleotide BLASTn search in the GenBank database. Based on the similarity search and taxonomic hierarchy, 10 reference sequences of 5 Myrophinae species and 34 sequences of 19 Ophichthinae species (family Ophichthidae) were retrieved from the GenBank database. The generated and acquired sequences were aligned by ClustalX program (Thompson et al. Citation1997) to make a comprehensive dataset with equal length and common start position. The mean genetic divergences were calculated using the Kimura 2 parameter (K2P) by MEGA6.0 (Tamura et al. Citation2013). Phylogenetic analysis was performed under the optimality criteria of Neighbor-Joining (NJ) by using PAUP* 4.0b10 (Swofford Citation2002) with 1000 bootstrap support. A sequence of Anguilla rostrata (Family Anguillidae) was also retrieved from GenBank and used as out-group in the dataset.
Result and discussion
During the survey at the outside of Nalban Bird Sanctuary, the luxurious growth of sea grass (Halophila sp.) was observed in Chilika lagoon. A total of 10 specimens of Anguilliformes were collected and identified as B. deraniyagalai, which is an addition to the Ichthyofauna of Chilika lagoon. This species was originally described from five specimens, collected from Arasalar River, Karaikkal, Tamil Nadu and the type specimen was deposited with the Zoological Survey of India, Kolkata (Menon Citation1961). The present study adopted both morphological and molecular data to validate the species identity and examined the existing genetic variation between the mentioned two morpho-groups hereunder.
Morphological data
The studied B. deraniyagalai was showed below mentioned morphological characters, body cylindrical, elongated with anus slightly before mid-body (preanal length 2.1–2.2 in total length). Dorsal fin origin on the head just before the gill opening and far behind to the rictus. Teeth small, slender, and blunt tipped. No premaxillary teeth, uniserial on maxilla and dentary. Two uniserial prevomerine teeth, vomerine teeth uniserial but placed irregularly. Pectoral fin reduced to a minute flap. There exist clearly two vertebral groups of the species: the first one with 5/81-83/190-196 and the second group with 5/72-74/177-178. Colour olive brown dorsally and dull yellow ventrally with a dark spot on ventral surface of the body, situated nearly one head length from gill opening.
Body notably elongated and almost cylindrical (). Depth at gill opening and anus nearly equal 66.4–88.2 in total length. Anus at about little before the midpoint of body with preanal length 2.1–2.2 in total length. Head about 15.0–19.8 in total length. Dorsal fin origin on head, just before gill opening and far behind the rictus; predorsal distance about 20.4–28.6 in total length. Greatest depth 3.81–5.5; snout 5.6–8.3; predorsal length 1.2–1.5 in head length. Eye small, located slightly closer to rictus than tip of snout, its diameter 3–4 in snout length. Upper jaw slightly longer than the lower jaw; upper jaw about 4.1–5 in head length and lower jaw about 5.0–6.6 in head length. Anterior nostril tubular, not reaching tip of snout; posterior nostril flap like open in upper lip. Gill opening below lateral line. Head pores are depicted as in . Dorsal fin and anal fin extending towards tip of tail but not confluent around tail, leaving tail tip exposed and hard (characteristic of subfamily Ophichthinae). Pectoral fin reduced to a minute flap, restricted to upper part of gill opening and difficult to distinguish. Lateral line pores 73–84 before vent and 10 or 11 before branchial opening (wide range more closely related to dimorphism in vertebral count). Teeth small, slender and blunt tipped. Premaxillary teeth absent; teeth uniserial on maxilla and dentary. Two uniserial prevomerine teeth present; vomerine teeth uniserial but placed irregularly ().
Figure 1. (A) Whole-body specimen of B. deraniyagalai collected from Chilika lagoon. (B) NJ tree of the studied eel species with bootstrap support. A. rostrata of Family Anguillidaeis used as an out-group in the phylogeny. Green and orange bars represent the two known subfamilies of family Ophichthidae. Two morpho-groups of studied B. deraniyagalai were showed by pink and blue voucher ID and accession numbers. (C) Line diagram of B. deraniyagalai. (D) Dental pattern of B. deraniyagalai.
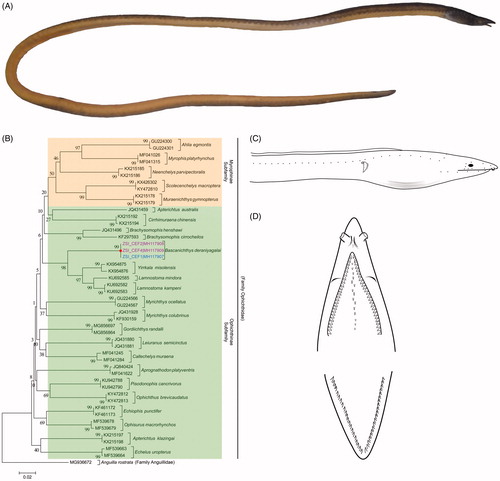
There exist clearly two vertebral groups of the species: the first one with five predorsal vertebrae, 81–83 preanal vertebrae and 190–196 total vertebrae (ZSI_CEF1) and the second group with five predorsal vertebrae, 72–74 preanal vertebrae and 176–178 total vertebrae (ZSI_CEF2 and ZSI_CEF4). The holotype is having the vertebrae 5/82/190. Colour-Body olive brown dorsally and dull yellow ventrally. A dark spot on ventral surface of the body; situated nearly one head length from gill opening. Lateral line pores pale and distinct in preserved specimens. The sexual dimorphism has been detected in Moringua species (Anguilliformes, Moringuidae), in which the male and female have different vertebral counts. However, the differences in vertebrae are uncommon in the members of Ophichthidae so far. To review the fact, two more fresh specimens were examined from the same group with higher vertebral (5/81-83/190-196), which were with eggs thus confirmed females. Thus, we confirmed that the specimen (ZSI_CEF1) with higher vertebral count was female. However, the sex determination could not confirm for the specimens (ZSI_CEF2 and ZSI_CEF4) with low vertebral count (5/72-74/177-178) due to long preservation in alcohol. Hence, the present study suggested there might be sexual dimorphism in B. deraniyagalai, which need further investigation with more sampling from different geographical regions.
Apart from the type locality, Arasalar river mouth (Menon Citation1961), this species has been reported from Godavari estuary (Krishnan and Mishra Citation2001), Krishna estuary (Mishra Citation2008), both Vamsadhara and Nagavali estuaries (Mishra Citation2010) of Andhra Pradesh. The specimens from Mahanadi estuary examined by one author (SSM) included in the report from Odisha coast (Barman et al. Citation2007) and record of Callechelys longipinnis from Rushikulya estuary (Rama Rao et al. Citation1992), Odisha found to be erroneous and referred here as this species. Hence, B. deraniyagalai is now confirmed to have a wider range of distribution along the east coast of India in almost all estuaries including Chilika Lake. The record of B. longipinnis from Sri Lanka may prove to be this species as observed by Menon (Citation1961).
Molecular data
The five species of Myrophinae and 20 species of Ophichthinae, including the studied species B. deraniyagalai, shows distinct clustering in the NJ phylogeny (). The two morpho-groups, ZSI_CEF1 (MH117907) and ZSI_CEF2, ZSI_CEF4 (MH117908, MH117909) depicts cohesive clustering and cladded as sister species of Yirrkala misolensis, Lamnostoma mindora, and Lamnostoma kampeni in the NJ tree. The overall mean genetic divergence of the eel data set was 17.8%. The within-species genetic divergence of the studied dataset was revealed ranging from 0% to 1.09%. Further, the genetic divergence among the species of Ophichthinae subfamily ranging from 3.1% to 21.6% with mean genetic divergence 16.5%. The genetic divergence among the species of Myrophinae subfamily ranging from 11.9% to 27.7% with mean genetic divergence 15.9%. The mean genetic divergence between two subfamilies of Ophichthidae resulted 20.4% in the current dataset. The B. deraniyagalai shows 0% K2P genetic divergence between two morpho-groups. The studied species was morphologically confused with the Lamnostoma species; however, sufficient genetic divergence (9.9–10.9%) was observed (). Based on both morphology and molecular data, the study effectively identified B. deraniyagalai species from Chilika Lake. Earlier this species might have been reported as Lamnostoma orientalis from Chilika lagoon as after extensive survey of two years no specimen of Lamnostoma orientalis have been collected.
Table 1. The between and within-species K2P genetic distance of the studied eel dataset.
The genus Bascanichthys comprises 17 species worldwide (Eschmeyer and Fong Citation2018). All these species are compared following original descriptions, as it was not possible to access type specimens at present. Of these, B. bascanium have pectoral fin as long as snout; B. bascanoides with pectoral slightly longer than eye; B. gaira with pectoral fin three to six times in its base and B. paulensis with a small, rounded pectoral fin, while in B. deraniyagalai, like rest of the species pectoral fin extremely reduced. B. ceciliae and B. inopinatus have higher vertebral count (225–226 and 198–205) and B. scuticaris with lesser count (159–167), verses 176–196 in B. deraniyagalei. B. pusillus is distinct in having biserial maxillary and vomerine teeth (uniserial in B. deraniyagalei); B. myersi and B. ceciliae have three to five intermaxillary teeth (absent in B. deraniyagalei). Tail length is distinctly shorter than the preanal length in B. cylindricus and B. panamensis. Bascanichthys sibogae can be distinguished in having gill opening in the dark area and three small conical papillae on upper jaw-one between nostrils, two behind nostrils. Only four species, B. fijiensis, B. filaria, B. kirkiiand B. longipinnis, are very closely similar to B. deraniyagalei. However, as observed by Menon (Citation1961), all these four species have anus about midpoint of body but B. deraniyagalaiis having anus nearer to snout than the tip of the tail. Head length of B. deraniyagalaiis longer than other four species (8.0–10.0 times in trunk versus 6.2–8.0 in B. deraniyagalei). Additionally, B. kirkii and B. longipinnis have a stouter body, body-depth about 3 times in head length (versus body depth 3.8–5.5 in the head for B. deraniyagalai). Further, B. filaria has biserial posterior maxillary teeth and pectoral fin developed into a minute filament. Although the taxonomic characters are available previously for this species, the present study added new morphometric characters, and variations recorded within B. deraniyagalai population in the same eco-system along with vertebral or sexual dimorphism. These data might be helpful for further taxonomic research on the particular group and helps to reduce the species misidentification. Further, a wide range of survey of the same and related taxa and generation of more nucleo-mitochondrial molecular data would render more authentic scenario of eel phylogeny and systematics research. This study also reports the species for the first time from Chilika lagoon.
Acknowledgements
We thank the Director, Zoological Survey of India, Ministry of Environment, Forest and Climate Change (MoEF&CC) for providing necessary working facilities.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Barman RP, Mishra SS, Kar S, Mukherjee P, Saren SC. 2007. Marine and estuarine fish fauna of Orissa. Rec Zool Surv India Occ Paper. 260:1–186.
- Bohlke EB. 1982. Vertebral formulae of type specimens of eels (Pisces: Anguilliformes). Proc Acad Nat Sci Phila. 134:31–49.
- Eschmeyer WN, Fong JD. 2018. Species by family/subfamily; [accessed 25 April 2018]. http://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp.
- Jamandre BWD, Shen KN, Yambot AV, Tzeng WN. 2007. Molecular phylogeny of Philippine freshwater eels Anguilla spp. (Actinopterygi: Anguilliformes: Anguillidae) inferred from mitochondrial DNA. Raffles B Zool. 14:51–59.
- Krishnan S, Mishra SS. 2001. Fauna of Godavari estuary. Fishes estuarine ecosystem series. Vol. 4. Kolkata (India): Zoological Survey of India.
- Laskar BA, Kumar V, Kundu S, Tyagi K, Chandra K. 2018. Taxonomic quest: validating two mahseer fishes (Actinopterygii: Cyprinidae) through molecular and morphological data from biodiversity hotspots in India. Hydrobiologia. 815:113–124.
- McCosker JE. 2010. Deepwater Indo-Pacific species of the snake-eel genus Ophichthus (Anguilliformes: Ophichthidae), with the description of nine new species. Zootaxa. 2505:1–39.
- Menon M. 1961. On a collection of fish from lagoon Chilika, Orissa. Rec Ind Mus. 59:41–69.
- Mishra SS. 2008. Fauna of Krishna Estuary. Fishes Estuarine Ecosystem Series. Vol. 5. Kolkata (India): Zoological Survey of India.
- Mishra SS. 2010. Fauna of Vamsadhara and Nagavali estuaries, Andhra Pradesh. Fishes Estuarine Ecosystem Series. Vol. 6. Kolkata (India): Zoological Survey of India.
- Mohapatra A, Ray D, Kumar V. 2013. A new fish species of the Genus Hapalogenys (Perciformes: Hapalogenyidae) from the Bay of Bengal, India. Zootaxa. 3718:367–377.
- Mohapatra A, Ray D, Smith DG, Mishra SS. 2016. A new species of elongate unpatterned moray eel of the genus Gymnothorax (Muraenidae: Muraeninae) from the Bay of Bengal. Zootaxa. 4150:591–598.
- Mohapatra A, Smith DG, Mohanty SR, Mishra SS, Tudu PC. 2017a. Gymnothorax visakhaensis sp. nov., a new species of elongate unpatterned moray eel (Muraenidae: Muraeninae) from the Indian Coast. Zootaxa. 4300:279–286.
- Mohapatra A, Smith DG, Mohanty SR, Mishra SS, Tudu PC. 2017b. Enchelycore propinqua sp. nov., a new moray eel (Anguilliformes: Muraenidae: Muraeninae) from the Indian Coast. Zootaxa. 4319:595–599.
- Mohapatra A, Smith DG, Ray D, Mishra SS, Mohanty SR. 2017c. Gymnothorax pseudotile sp. nov. (Muraenidae: Muraeninae) from Bay of Bengal India. Zootaxa. 4286:586–592.
- Peninal S, Subramanian J, Elavarasi A, Kalaiselvam M. 2017. Genetic identification of marine eels through DNA barcoding from Parangipettai coastal waters. Genomics Data. 11:81–84.
- Rama Rao KV, NageswaraRao CA, Nahar SC, Rao DV, Mohapatra A. 1992. Studies on the ecology and fauna of the Rushikulya estuary, (Ganjam), Orissa. Estuarine Ecosystem Series, Part 1: Rushikulya estuary, Orissa. Zool Surv India Kolkata. 7–26.
- Ray D, Mohapatra A, Smith DG. 2015. A new species of short brown unpatterned moray eel of the genus Gymnothorax (Anguilliformes: Muraenidae) from the Bay of Bengal. Zootaxa. 4027:140–144.
- Santini F, Kong X, Sorenson L, Carnevale G, Mehta RS, Alfaro ME. 2013. A multi-locus molecular timescale for the origin and diversification of eels (Order: Anguilliformes.). Mol Phylogenet Evol. 69(3):884–894.
- Swofford DL. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods).Version 4. Sunderland (MA): Sinauer Associates.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tang KL, Fielitz C. 2012. Phylogeny of moray eels (Anguilliformes: Muraenidae), with a revised classification of true eels (Teleostei: Elopomorpha: Anguilliformes). Mitochondrial DNA. 24(1):55–66.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882.
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert P. 2005. DNA barcoding Australia's fish species. Philos Trans R Soc Lond B Biol Sci. 360:1847–1857.
