Abstract
The Odontobutis potamophila is endemic commercial fishery species to China and distributed mainly in middle and lower Yangtze River of China. Here, we describe a complete mitochondrial genome from the newly found population of O. potamophila in the Three Gorges Reservoir where located in the upper Yangtze River. It was 16,939 bp in size and encodes for two rRNAs, 22 tRNAs, 13 protein-coding genes, and a D-loop control region. Phylogenetic analysis confirmed the evolutionary relationship of the O. potamophila in the upper Yangtze to other five species of the genus Odontobutis. The mitochondrial genome would help to elucidate the origin of the new population of O. potamophila.
Odontobutis potamophila (Günther, 1861), belongs to the genus Odontobutis with high economic value in China, mainly distributes in the middle and lower reaches of the Yangtze River and its tributaries (Wu et al. Citation1993). The all reports regarding genetic characteristics of O. potamophila based on samples collected in the middle and lower reaches of the Yangtze River basin (Ma et al. Citation2015; Xu et al. Citation2015; Li and Liu Citation2016). Three Gorges Reservoir (TGR) was completely formed in 2006 after the Three Gorges Dam cut the upper Yangtze River. The original river flow pattern was changed in TGR, which caused alteration of the population structure, species composition, and spatial and temporal distributions of fishes (Xie et al. Citation2018). According to historical documents, there is no record about the distribution of O. potamophila in the upper Yangtze River basin (Ding Citation1994). However, we collected 20 specimens of O. potamophila in the TGR of the upper Yangtze River, Chongqing of China in 2018. To our knowledge, this is a newly found population of O. potamophila in the upper Yangtze River. The specimens were stored in 90% ethanol and kept in the fishes and animal specimens room (CAST 30-0119) of Museum of Southwest University.
In this study, we determined the complete mtDNA sequence of an O. potamophila (100.5 mm standard length, 13.50 g mass, No. 20180100011), which was collected from Jiulongpo (29.527382 N, 106.53606 E) in TGR of Chongqing during October 2018. Total genomic DNA extracted from dorsal muscle tissues by the standard procedure (Sambrook and Russell Citation2001). Twenty-seven pairs of PCR primers were designed to amplify the segments using PCR and then products were sequenced and assembled. The mitochondrial genome of O. potamophila (GenBank accession No. MK408452) was a circular molecule of 16,939 bp with 13 protein-coding genes, two rRNA, 22 tRNA genes, and D-Loop region of 859 bp. All genes show the typical gene arrangement conforming to the vertebrate consensus (Noack et al. Citation1996). Except for the eight tRNA genes and ND6 gene, all the other mitochondrial genes were encoded on the heavy strand, which was similar to other fishes (Hu et al. Citation2015). There were two overlapping sequences between genes. Among the 13 protein-coding genes, except COX2 and CYTB use an incomplete stop codon ‘T,’ the rest are encoded by the typical TAA or TAG stop codons and except COX1 with GTG as start codon, the remaining 11 protein-coding genes start with an ATG codon.
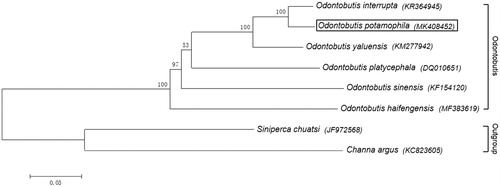
The sequences of complete mitochondrial genome of O. potamophila and other 7 species including 5 species from the genus Odontobutis and 2 species as outgroup were used for phylogenetic analysis (). The results confirmed that O. potamophila is closely related to the O. interrupta than other 4 species in the genus Odontobutis, within a monophyletic clade (Ma et al. Citation2015). The mitogenome would help to elucidate the origin of the new upstream population of O. potamophila.
Figure 1. Phylogenetic tree of six species of the genus Odontobutis based on the sequences of complete mitochondrial genome by Bayesian inference and maximum likelihood methods. Siniperca chuatsi from Family Serranidae, Channa argus from Family Channidae as outgroup. Phylogenetic analysis confirms that O. potamophila (indicated as boxed) belongs to the clade of the genus Odontobutis and more closely related to the O. interrupta. Bootstrap values (1000 replicates) are shown for each node, and GenBank accession numbers are provided in parentheses accompanying each taxon.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Ding RH. 1994. The fishes of Sichuan. Chengdu, China: Sichuan Publishing House of Science and Technology.
- Hu YX, Zhou Q, Song Y, Chen DQ, Li Y. 2015. Complete mitochondrial genome of the smallscale yellowfin, Plagiognathops microlepis (Teleostei: Cypriniformes: Cyprinidae). Mitochondr DNA. 26:463–464.
- Li Q, Liu ZZ. 2016. New complete mitochondrial genome of the Odontobutis potamophila (Perciformes, Odontobutidae): genome description and phylogenetic performance. Mitochondrial DNA A. 27:163–164.
- Ma ZH, Yang XF, Bercsenyi M, Wu JJ, Yu YY, Wei KJ, Fan QX, Yang RB. 2015. Comparative mitogenomics of the genus Odontobutis (Perciformes: Gobioidei: Odontobutidae) revealed conserved gene rearrangement and high sequence variations. IJMS. 16:25031–25049.
- Noack K, Zardoya R, Meyer A. 1996. The complete mitochondrial DNA sequence of the bichir (Polypterus ornatipinnis), a basal ray-finned fish: ancient establishment of the consensus vertebrate gene order. Genetics. 144:1165–1180.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. 3rd ed. Beijing, China: Science Press.
- Wu HL, Wu XQ, Xie YH. 1993. A revision of the genus Odontobutis from China with description of a new species. J Shanghai Fish Univ. 2:52–61.
- Xie CY, Niu YB, Luo DH, Feng XW, Pu DY, Peng ZG, Zeng B, Wang ZJ. 2018. A preliminary study on fish diversity for some important branches of Three Gorges Reservoir. Resour Env Yangtze Basin. 27:2747–2756.
- Xu Y, Zhong LQ, Li XX, Zhu XH, Wang CH, Shi YB. 2015. Genetic variation analysis of Odontobutis potamophila from five lakes based on mitochondrial DNA cyt b. J Lake Sci. 27:693–699.
