Abstract
The complete mitochondrial genome of Sardinella fijiensis was determined in this study, which was 16,651bp long, containing 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, a putative control region and one origin of replication on the light-strand. The overall base composition includes C (29.4%), A (26.4%), T (24.7%), and G (19.5%). Moreover, the 13 PCGs encode 3799 amino acids in total, 12 of which use the initiation codon ATG except COI uses GTG, most of them have complete stop codon, whereas three protein-coding genes (COII, ND4, and Cytb) ended with the incomplete stop codon represented as a single T. The phylogenetic tree based on the Neighbor Joining method was constructed to provide relationship within Clupeidae, which could be a useful basis for the further understanding of this species.
Sardinella fijiensis is a species of ray-finned fish in the genus Sardinella, which found in the Atlantic, Indian, and the Pacific Ocean (Sektiana et al. Citation2017). S. fijiensis is important not only in marine food webs as a base consumer supporting tuna, seabirds, and marine mammals (Willette et al. Citation2011). Considering its ecological importance and commercial value, to gain its molecular information, we described the complete mitogenome of S. fijiensis, and explored the phylogenetic relationship within Percoidea, the available complete mitogenome was used to provide additional reference to not only the phylogenetic relationship of S. fijiensis and related species, but also understanding for gene rearrangements and eel evolution in the further.
The specimen was sampled by commercial bottom trawling in the South China Sea (16°48′48″N; 112°22′12″E) and stored in laboratory of Second Institute of Oceanography with accession number FJ001. Total genomic DNA was extracted from muscle tissue of individual using the phenol-chloroform method (Köchl et al. Citation2005).
Similar to the typical mitogenome of vertebrates, the mitogenome of S. fijiensis is a closed double-stranded circular molecule of 16,651bp (GenBank accession No. MK400441), which contains 13 protein-coding genes, 2 ribosomal RNA genes, 22 tRNA genes, and 2 main non-coding regions (Miya et al. Citation2001). The overall base composition is 26.4%%, 24.7%, 29.4% and 19.5% for A, T, C, and G, respectively, with a slight AT bias (51.1%). Most mitochondrial genes are encoded on the heavy strand except for ND6 and eight tRNA genes (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, Pro), which are encoded on the light strand (Zeng et al. Citation2012). 13 PCGs encode 3799 amino acids in total, all of them use the initiation codon ATG except COI uses GTG, which is quite common in vertebrate mtDNA (Zhu, Gong, Jiang, et al. Citation2018; Zhu, Lü, et al. Citation2018). Most of them have TAA or TAG as the complete stop codon, whereas three protein-coding genes (COII, ND4, and Cytb) ended with an incomplete stop codon T. The 12S rRNA and 16S rRNA are 951 and 1686bp, which separated by tRNA-Val. the tRNA genes were generated using the program tRNAs-can-SE (Lowe and Eddy Citation1997), all of them can fold into a typical cloverleaf structure except tRNA-Ser (AGY), which lacks a dihydrouridine arm (Ojala et al. Citation1981). The CR is determined to be 985bp, by comparing the sequences of the CR with other teleost, three typical domains are observed, including termination-associated sequences, the central conserved sequence block domain and the conserved sequence block domain, which is identical to that in other teleostean mitogenomes (Gong et al. Citation2017; Zhu, Gong, Lü, et al. Citation2018).
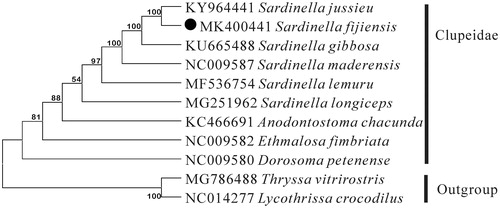
To explore the phylogenetic position of S. fijiensis, a phylogenetic tree was constructed based on the Neighbor Joining (NJ) analysis of 12 PCGs encoded by the heavy strand. The results of the present study supports S. fijiensis has a closest relationship with Sardinella jussieu, highly supported by a bootstrap value of 100 (), The results in this study could be a useful basis for better understanding of this species and were expected to provide additional reference to the phylogenetic relationship of S. fijiensis and related species.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Gong L, Liu LQ, Guo BY, Ye YY, Lü ZM. 2017. The complete mitochondrial genome characterization of Thunnus obesus (Scombriformes: Scombridae) and phylogenetic analyses of Thunnus. Conserv Genet Resour. 9:1–5.
- Köchl S, Niederstätter H, Parson W. 2005. DNA extraction and quantitation of forensic samples using the phenol-chloroform method and real-time PCR. Methods Mol Biol. 297:13.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18:1993–2009.
- Ojala D, Montoya J, Attardi G. 1981. TRNA punctuation model of RNA processing in human mitochondrial. Nature. 290:470–474.
- Sektiana SP, Andriyono S, Kim H-W. 2017. Characterization of the complete mitochondrial genome of Mauritian sardinella, Sardinella jussieu (Lacepède, 1803), collected in the Banten Bay, Indonesia. Fish Aquat Sci. 20:26.
- Willette DA, Santos MD, Aragon MA. 2011. First report of the Taiwan sardinella Sardinella hualiensis (Clupeiformes: Clupeidae) in the Philippines. J Fish Biol. 79:2087–2094.
- Zeng Z, Liu ZZ, Pan LD, Tang SJ, Wang CT, Tang WQ, Yang JQ. 2012. Complete mitochondrial genome of the endangered roughskin sculpin Trachidermus fasciatus (Scorpaeniformes, Cottidae). Mitochondrial DNA. 23:435–437.
- Zhu K, Gong L, Jiang L, Liu L, Lü Z, Liu B-J. 2018. Phylogenetic analysis of the complete mitochondrial genome of Anguilla japonica (Anguilliformes, Anguillidae). Mitochondrial DNA Part B. 3:536–537.
- Zhu K, Gong L, Lü Z, Liu L, Jiang L, Liu B. 2018. The complete mitochondrial genome of Chaetodon octofasciatus (Perciformes: Chaetodontidae) and phylogenetic studies of Percoidea. Mitochondrial DNA Part B. 3:531–532.
- Zhu K, Lü Z, Liu L, Gong L, Liu B. 2018. The complete mitochondrial genome of Trachidermus fasciatus (Scorpaeniformes: Cottidae) and phylogenetic studies of Cottidae. Mitochondrial DNA Part B. 3:301–302.