Abstract
Epophthalmia elegans is one of edible aquatic insects in Southwest China. In this study, the complete mitochondrial genome E. elegans (Odonata, Corduliidae) was determined. The mitogenome is 15,719 bp in size (GenBank accession number MK522522), including 13 protein-coding (PCGs), 22 transfer RNAs, 2 ribosomal RNAs genes, and a noncoding D-loop region. The overall base composition of E. elegans mitogenome is 40.40% for A, 15.09% for C, 33.18 for T, and 11.33% for G, with a high AT bias of 73.58%. This mitogenome data can contribute to our understanding of the phylogeny and evolution of E. elegans.
Dragonflies are good models to study insect ecology and evolution (Bybee et al. Citation2016). Epophthalmia elegans (Brauer 1865) is a species of big dragonfly, mainly distributed in Indomalaya (Southern China and Philippines) and Eastern Palaearctic (Tennessen Citation1997). The nymphs of this dragonfly are popular as food in Southwest China. Elucidating the structure of E. elegans mitogenome is important for understanding its diversity and evolution.
The specimen of E. elegans used in this study was collected from Yongsheng County, Yunnan Province, China (N 25°59′, E 100°22′) and deposited in the insect specimen room of Research Institute of Resource Insects with an accession number RIRI-w-20181024. Total mitochondrial genomic DNA was sequenced by the method of Sanger dideoxy. According to the mtDNA sequences, protein-coding genes (PCGs) were identified using BLAST search in NCBI, and tRNA genes were identified using the tRNAscan-SE search server (Schattner et al. Citation2005).
The complete mitogenome of E. elegans is 15,719 bp in size (GenBank accession number MK522522), with a GC content of 26.42%, showing an obvious AT mutation bias (Eyre-Walker Citation1997). The D-loop region exhibits the highest A + T content (91.92%) in the E. elegans mitogenome. This region has been shown to harbour the origin sites for the transcription and replication for both strands of insect mitogenomes (Yukuhiro et al. Citation2002). The gene order and orientation are identical to the most common type suggested as ancestral for insects (Cameron Citation2014). All the 13 PCGs use standard ATN (Met) start codons. All the tRNAs except tRNASer(AGN) could be folded into the typical cloverleaf secondary structures. The unusual tRNASer(AGN) lacks dihydrouridine (DHU) arm.
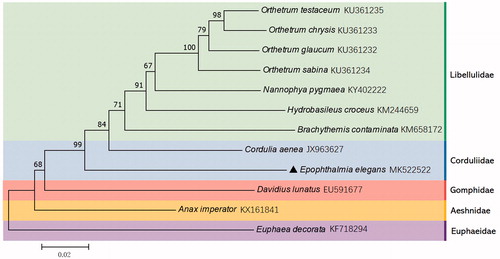
Based on the concatenated amino acid sequences of 13 PCGs, the neighbour-joining method was used to construct the phylogenetic relationship of E. elegans with 11 other dragonflies (). The result showed that E. elegans is closely related to Cordulia aenea (Odonata: Corduliidae), which is in accordance with the traditional morphological classification and recent molecular research (Carle et al. Citation2015). The present data could contribute to detailed phylogeography and other related analysis of E. elegans.
Figure 1. Phylogenetic tree showing the relationship between E. elegans and 11 other dragonflies based on neighbour-joining method. Euphaea decorata (Odonata: Zygoptera) was used as an outgroup relative to the other 11 individuals (Odonata: Anisoptera). GenBank accession numbers of each species were listed in the tree.
Disclosure statement
The authors declare no competing materials in the preparation and execution of this manuscript. The authors are responsible for the content and writing of this article.
Additional information
Funding
References
- Bybee S, Córdoba-Aguilar A, Duryea MC, Futahashi R, Hansson B, Lorenzo-Carballa MO, Schilder R, Stoks R, Suvorov A, Svensson EI, et al. 2016. Odonata (dragonflies and damselflies) as a bridge between ecology and evolutionary genomics. Front Zool. 13:46.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Carle FL, Kjer KM, May ML. 2015. A molecular phylogeny and classification of Anisoptera (Odonata.). Arthropod Syst Phylog. 73:281–301.
- Eyre-Walker A. 1997. Differentiating between selection and mutation bias. Genetics. 147:1983–1987.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686.
- Tennessen JK. 1997. The rate of species descriptions in Odonata. Entomol News. 108:122–126.
- Yukuhiro K, Sezutsu H, Itoh M, Shimizu K, Banno Y. 2002. Significant levels of sequence divergence and gene rearrangements have occurred between the mitochondrial genomes of the wild mulberry silkmoth, Bombyx mandarina, and its close relative, the domesticated silkmoth, Bombyx mori. Mol Biol Evol. 19:1385–1389.
