Abstract
The complete of mitochondrial genomes in two kinds of Corbicula fluminea, black and yellow, are 17,293 bp and 17,068 bp in length, respectively. Both of them contain 13 protein-coding genes (PCGs), two ribosomal RNA genes, 22 transfer RNA genes (tRNAs), and one control region (CRs). The proportion of base-pairs in two kinds of C. fluminea are different, with A + T (68.78%) and G + C (30.17%) in black one as well as A + T (70.39%) and G + C (29.61%) in yellow one. Besides, the secondary structures in two kinds of clam, including trnS2, trnD, trnE, trnI, trnL1 and trnT, are various. This paper demonstrates the range of variation in mtDNA which could explore the relationship between the C. fluminea and geographical distribution.
Corbicula fluminea (C. fluminea) is native to Asia and then spread to North America, South America, and Europe. Corbicula fluminea has always been popular among the people because of its rich nutrition and pharmaceutical value, and it is one of the essential fishery objects in China. In recent years, with the popularity of C. fluminea in China, Japan, and Southeast Asia, the breeding scale of C. fluminea has gradually expanded in freshwater aquaculture, especially the artificial breeding. Corbicula fluminea has two different shell colours, black (C. fluminea B) and yellow (C. fluminea Y). Like other vertebrates, the mitochondrion genomes of this kind of shellfish are composed of 13 protein-coding genes, 22 tRNAs, and two rRNA genes (Han et al. Citation2016; He et al. Citation2016; Wang et al. Citation2016).
The specimens were collected from the Hongze Lake, Suqian county, Jiangsu Province, China (North latitude 33°08′―33°44′, East longitude 117°56′―118°46′) where 100 samples from two kinds of C. fluminea were collected separately. The qualified extracted mitochondrial DNAs were preserved at −20 °C in Key Laboratory of Genetic Resources for Freshwater Aquaculture and Fisheries, Ministry of Agriculture, Shanghai Ocean University.
The complete mitochondrial genome of C. fluminea B is 17,293 bp in length, whereas C. fluminea Y is 17,068 bp in length. The transfer RNA genes and ribosomal RNA genes of the C. fluminea Y mitochondrial genome are 1409 and 2107 nucleotides long, respectively. A total of 22 tRNA sequences were found throughout the mitogenome of C. fluminea Y, which identified based on their respective anticodons and secondary structures, ranged in length from 62 (trnR, trnH, trnQ, trnW) to 68 bp (trnM). Two ribosomal RNA genes (12s rRNA and 16s rRNA gene) were found throughout the C. fluminea Y mitogenome. 12s rRNA (rrnS) gene is 869 bp in length, which lies between the trnT and trnM genes, whereas 16s rRNA (rrnL) gene is 1238 bp in length lying between the cob and atp8 genes. The A + T content of two rRNAs are 66.51% and 73.51%, respectively, which are higher than C. fluminea B. The AT skew of rrnL is negative (−0.015) while the GC skew of rrnL (0.293) and AT skew (0.042), GC skew (0.292) of rrnS are positive that it is the same as C. fluminea B. All the tRNAs have a standard cloverleaf structure, except for trnS2 with a missing dihydorouridine arm (C. fluminea B) and dihydorouridine loop (C. fluminea Y).
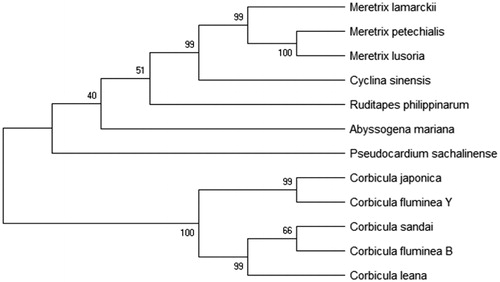
In general, C. fluminea B and C. fluminea Y were first determined in this research. Corbicula fluminea B relative is the closest to the C. sandai and C. fluminea Y is the closest to the C. japonica. The range of variation of mtDNA is closely related to habitat in order that the genetic variability of freshwater animal is often greater than seawater’s. The study of the relationship between mtDNA and geographical distribution found that mtDNA with close genetic relationship is geographically continuous as well.
Disclosure statement
No potential conflict of interest was reported by the authors.
Figure 1. Phylogenetic trees were constructed by MEGA5.1 software to analyze the phylogenetic relationship between C. fluminea B and C. fluminea Y with Maximum Parsimony, Neighbor-Joining, Maximum Likelihood methods, and 12 mitogenome used, including Corbicula japonica (BAO96500), Corbicula sandai (BAO96595), Corbicula leana (BAP25514), Meretrix lamarckii (AKE36664), Pseudocardium sachalinense (ATZ68907), Meretrix petechialis (YP002929378), Meretrix lusoria (YP004072670), Meretrix lamarckii (YP004934874), Cyclina sinensis (YP009236122), Ruditapes philippinarum (YP009305271), C. fluminea B(MK 587518), and C. fluminea Y(MK 587517).
Additional information
Funding
References
- Han Z, Wang G, Xue T. 2016. The F-type complete mitochondrial genome of Chinese freshwater mussels Cuneopsis pisciculus [J]. Mitochondrial DNA. 27:3376–3377.
- He F, Wang G, Li LJ. 2016. Complete F-type mitochondrial genome of Chinese freshwater mussels Lamprotula gottschei. Mitochondrial DNA A DNA Mapp Seq Anal. 27:246–247.
- Wang G, Chen M, Li J. 2016. Complete F-type mitochondrial genome of freshwater mussel Lanceolaria glayana. Mitochondrial DNA A DNA Mapp Seq Anal. 27:846–847.
