Abstract
Japanese Millet, Echinochloa esculenta is a weed-suppressing cover crop that was grown once as a staple food crop in Japanese regions when rice cultivation failed. Here, we report the complete chloroplast (cp) genome sequence of E. esculenta for the first time to understand the diversification of this species in the family Poaceae. The size of the circular chloroplast genome is 139,851 bp in length with 38.6% overall GC content which exhibits a typical quadripartite structure, containing pair of inverted repeats of 22,748 bp, flanked by large single copy and small single copy regions of 81,837 bp and 12,518 bp, respectively. The cp genome encodes 111 unique genes, 76 of which are protein-coding genes, four rRNA genes, 30 tRNA genes, and 18 duplicated genes in the inverted repeat region. The phylogenetic analysis indicated that the E. esculenta is closely related with the wild relative barnyard grass (E. crus-galli).
Introduction
Japanese millet (Echinochloa esculenta), formerly known as E. utilis, is an annual, warm-season grass native to eastern Asia. It is grown primarily as forage and cultivated mostly in the temperate regions of Japan, Korea, China, Russia, and Germany (De Wet et al. Citation1983). It is also believed to have as a domesticated species directly derived from the wild millet barnyard grass (E. crus-galli) some 4000 years ago in Japan (Doggett Citation1989). In addition to this domesticated species, Echinochloa includes about 20–30 annual and perennial wild species distributed throughout the warmer parts of the world (Clayton and Renvoize Citation1986). In general, inter-specific hybridization between the species promises with the long-term view of mutual introgression of desirable genes between the species, which has enormous potential for their mutual genetic enhancement.
Several morphological traits have been used for finding the species relationships but they do not precisely describe the genetic relationship, due to various environmental influences. Considering the genetic erosion, understanding the relationships between species is vital for efficient inter-specific hybridization to transfer of valuable traits from wild species to cultivar germplasm. Chloroplast genome sequencing has been extensively applied to reveal the species evolution and diversity (Daniell et al. Citation2016; Liu et al. Citation2016; Park Citation2017). Chloroplast genome sequence of Echinochloa species have been reported (Ye et al. Citation2014; Hereward et al. Citation2016; Perumal et al. Citation2016). Here, we report the chloroplast genome of E. esculenta to find its internal relationships within the family Poaceae.
Japanese millet (Accession No: IT230633) was obtained from the national genebank division of RDA, Republic of Korea. Genomic DNA of the 40-day-old seedling was used to sequence the genome by Illumina MiSeq instrument with pair-end library (2 × 300 bp) at LabGenomics (http://www.Lab.genomics.com/kor/). A total of 5,118,176 raw reads and 3,409,059 clean reads were obtained and mapped with the reference cp genome, A. sativa L. (GenBank accession KM974733), which contains 226,500 aligned reads with about average 352× coverage. The cp genome was annotated using DOGMA (http://dogma.ccbb.utexas.edu/) software and tRNAscan-SE (Lowe and Chan Citation2016).
The chloroplast genome has a total length of 139, 851 bp with 38.6% overall GC content (NCBI accession number KX756178) which exhibit characteristic quadripartite structure, consisting of LSC region of 81,837 bp, two IR regions of 22,748 bp and SSC region of 12,518 bp. The chloroplast genome was predicted to encode 111 genes, including 76 protein-coding genes, 30 tRNA genes, four rRNA genes. A total of 18 genes were duplicated in the inverted repeat regions, eight genes, and two tRNA genes contain one intron while ycf3 has two introns.
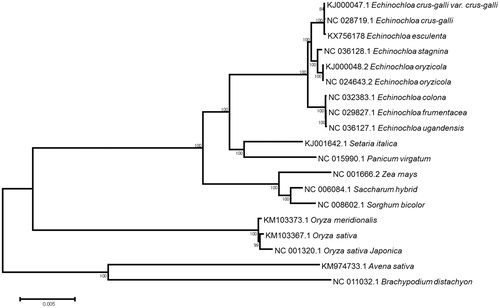
Phylogenetic analysis was performed based on the published chloroplast genome sequences from Poaceae species which were downloaded from the NCBI database. Whole genome sequence was aligned using MAFFT v7.304 (Katoh and Standley Citation2016), and MEGA6 (Tamura et al. Citation2013) software was used to construct a maximum likelihood (ML) tree with 1000 bootstrap replicates. Phylogenomic analyses of Poaceae cp genomes revealed that E. esculenta is clustered with its wild relative in the Echinochloa genus of the grass or poaceae family (). The cp genome of E. esculenta will provide a valuable resource for germplasm collection, conservation, and utilization.
Disclosure statement
The authors are highly grateful to the published genome data in the public database. The authors declare no conflicts of interest and are responsible for the content.
Additional information
Funding
References
- Clayton WD, Renvoize SA. 1986. Genera graminum. Kew Bull Add Ser. 13:280–281.
- Daniell H, Lin CS, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17:134
- De Wet JMJ, Rao KEP, Mengesha MH, Brink DE. 1983. Domestication of Sawa millet (Echinochloa colona). Economic Bot. 37:283–291.
- Doggett H. 1989. Small millets: a selective overview. In: Seetharam A, Riley K, Harinaryana G, editors. Small millets in global agriculture. New Delhi: Oxford & IBH; p. 3–18.
- Hereward JP, Werth JA, Thornby DF, Keenan M, Chauhan BS, Walter GH. 2016. Complete chloroplast genome sequences of six lines of Echinochloa colona (L.) link. Mitochondrial DNA B. 1:945–946.
- Katoh K, Standley DM. 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics (Oxford, England). 32:1933–1942.
- Liu F, Tembrock LR, Sun C, Han G, Guo C, Wu Z. 2016. The complete plastid genome of the wild rice species Oryza brachyantha (Poaceae). Mitochondrial DNA B. 1:218–219.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Park T-H. 2017. The complete chloroplast genome of Solanum berthaultii, one of the potato wild relative species. Mitochondrial DNA B. 2:88–89.
- Perumal S, Jayakodi M, Kim D-S, Yang T-j, Natesan S. 2016. The complete chloroplast genome sequence of Indian barnyard millet, Echinochloa frumentacea (Poaceae). Mitochondrial DNA B. 1:79–80.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Ye CY, Lin Z, Li G, Wang YY, Qiu J, Fu F, Zhang H, Chen L, Ye S, Song W, et al. 2014. Echinochloa chloroplast genomes: insights into the evolution and taxonomic identification of two weedy species. PLoS One. 9:e113657.