Abstract
Rhizophora × lamarckii is a natural hybrid of R. apiculata and R. stylosa, and it is also an endangered mangrove material in China with the exclusive location in Hainan Island. In this paper, we reported and characterized the complete chloroplast genome sequence of R. × lamarckii by assembling from short reads generated using Illumina sequencing. Total chloroplast genome size was 164,325 bp, including inverted repeats (IRs) of 26,325 bp, separated by a large single copy (LSC) and a small single copy (SSC) of 92,433 bp and 19,244 bp, respectively. A total of 121 genes, including 42 tRNAs, eight rRNAs, and 81 protein-coding genes, were identified. The GC content of R. × lamarckii is 34.67%. Phylogenetic analysis using the maximum likelihood algorithm showed that R. × lamarckii is sister to a clade with four species in the order Malpighiales.
Rhizophora × lamarckii Montrouz (Rhizophoraceae) is a natural hybrid mangrove material of R. apiculata and R. stylosa, which are the two important mangrove species of Rhizophora (Duke Citation2006; Luo et al. Citation2017). This natural hybrid with its parents is restricted to the Indo-West Pacific (IWP) region and it is only found in the hybridization zone of two parents (Tomlinson Citation1986; Duke Citation2010). This is an endangered mangrove material of sporadic occurrence with the abortion character, and it is identified in Hainan Island, China until 2017 (Luo et al. Citation2017). Now, research on R. × lamarckii has focused on its geographical distribution, morphometric characteristics and flowering phenology (Luo et al. Citation2017). The information of chloroplast genomes has been extensively applied to understanding plant genetic diversity and natural hybrid forming mechanism (Wang Citation2017). In this paper, we characterized the complete chloroplast genome sequence of R. × lamarckii to contribute for further phylogenetic and protective studies of this natural hybrid.
Fresh leaves were collected from six individual of R. × lamarckii in XinYing Mangrove Natural Garden, Hainan Island (N19°30', E109°30'), China. The total genomic DNA was extracted from ten mixed fresh leaves of R. × lamarckii by using the modified CTAB method (Doyle Citation1987) in the laboratory of Lingnan Normal University. Further, the specimen was stored in the herbarium of Lingnan Normal University. Genome sequencing was performed on an Illumina Hiseq X Ten platform with paired-end reads of 150 bp. In total, 68.8 Mb short sequence data with Q20 was 97.16% was obtained. The remaining high-quality reads were used to assemble the chloroplast genome in SOAPdenovo2 (Luo et al. Citation2012). The genes annotation in the chloroplast genome was doing with the DOGMA Program with manual check and the circular chloroplast genome map was drawn by using OGDRAW (Wyman et al. Citation2004). The accession number in Genbank is MK392466. Six chloroplast genome sequences were aligned by using the complete chloroplast genomes, five plants from Malpighiales including R. × lamarckii, and Oryza sativa was used as the out-group species. Phylogenetic analysis using the maximum likelihood algorithm was conducted with RAxML (Stamatakis Citation2014) implemented in Geneious ver. 10.1 (Kearse et al. Citation2012).
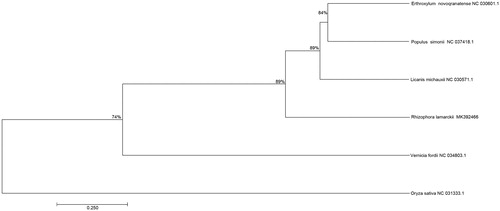
A typical quadripartite structure was found in the chloroplast genome of R. × lamarckii with a length of 164,325 bp, which contains inverted repeats (IRs) of 26,325 bp, separated by a large single copy (LSC) and a small single copy (SSC) of 92,433 bp and 19,244 bp, respectively. The chloroplast genome has a GC content of 34.67% and contains a total of 121 predicted genes, including 42 tRNAs, eight rRNAs, and 81 protein-coding genes. Among nine genes with intron, six protein-coding genes (rpoC1, rps20, atpF, rpl2, ndhA, and ndhB) contain one intron and three genes (petD, rps7, and ycf3) contain two introns. Phylogenetic analysis with other seven plant chloroplast genomes showed that R. × lamarckii is sister to a clade containing four species in Malpighiales (). The useful genomic resources for characterization of genetic diversity of R. × lamarckii by the chloroplast genome will help for the study of natural hybridization mechanism.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Duke N. 2006. Australia’s mangroves: the authoriative guide to australiàs mangrove plants. Brisbane, QLD: University of Queensland; p. 162–171.
- Duke N. 2010. Overlap of eastern and western mangroves in the South-western Pacific: hybridization of all three Rhizophora (Rhizophoraceae) combinations in New Cledonia. Blum – J Plant Tax and Plant Geog. 55:171–188.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Luo L, Zhong C, Hou X, Wang W. 2017. Rhizophora × lamarckii, a new recorded mangrove species in China. J Xiamen Univ (Nat Sci). 56:346–350.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tomlinson P. 1986. The botany of mangrove. Cambridge: Cambridge University Press; p. 317–360.
- Wang Y. 2017. Natrual hybridization and speciation. Biodivers Sci. 25:565–576.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.