Abstract
The mitochondrial genome of Geothelphusa sp. DJL-2014 isolate Gm01 (named Naju crab) was sequenced nearly completely and analyzed. It is 18052 bp in length, consisting 13 protein-coding genes (PCGs), two ribosomal RNA genes, 22 transfer RNA genes, and a control region. In Geothelphusa sp. DJL-2014 isolate Gm01, there are two trnLs, as shown in animal mtDNA. On the contrary, in Geothelphusa dehaani (AB187570), there are three trnLs.
The genus Geothelphusa (Stimpson, 1858) (Crustacea, Decapoda, Potamidae, Potamiscinae) is one of eight families of freshwater crab. It is endemic to the islands of Far East Asia, where the sixty-five Geothelphusa spp. reportedly inhabit (http://www.marinespecies.org/aphia.php?p=taxdetails&id=391198). The true freshwater crab has recently been reported in Korea (Lee and Kim Citation2019). A specimen of freshwater crabs was collected from mountain stream in Pungnim-ri, Nampyeong-eup, Naju city, Jeollanam-do, Republic of Korea (GIS: 35.0N 126.8E). The exact location is not given because this crab would be endangered. Genomic DNA of this crab can be found at Department of Biological Sciences, the Chonnam National University, under the certificate number DJL-2014 isolate Gm01. Genomic DNA extraction, nucleotide sequencing, gene annotation, and phylogenetic analyses were performed (Boore Citation1999; Liu and Cui Citation2010, Yeo et al. Citation2008).
There are 18,052 base pairs (bp) in the mitochondrial genome of Geothelphusa sp. DJL-2014 isolate Gm01 (GenBank accession no. MG674171), which was not completely sequenced due to long repetition nucleotide: its total length is about 24 kb. All genes of the standard metazoan mitochondrial genome are found using MITOS (v917), consisting 13 protein-coding genes (PCG), two ribosomal RNA genes (rRNA), 22 transfer RNA genes (tRNA), and a control region (Bernt et al. Citation2013). The Geothelphusa sp. DJL-2014 isolate Gm01 showed the same gene order of G. dehaani: The gene arrangement was identical to that of the compared G. dehaani except for trnL2cun (Segawa and Aotsuka Citation2005). The initial codon is the typical ATN, with the most common being ATG and another being ATA (APT6, ND1, ND3, ND5 and ND6. The most frequent complete termination codon is TAA in nine genes. ND1 and ND3 are terminated with TAG codon. CO2 and CYTB are terminated with the abbreviated stop codon T. The 22 tRNA genes vary from 62 to 73 bp. Two trnLs were found in Geothelphusa sp. DJL-2014 isolate Gm01, as in animal mtDNA, whereas three trnLs were found in G. dehaani. The control regions (CRs) are located between the rrnS and tRNAIle genes, as in most of the brachyuran mitochondrial genomes.
Eleven PCGs were aligned in ClustalW based on codons using the default settings. The phylogenetic position of Geothelphusa sp. DJL-2014 isolate Gm01 was determined on 11 PCGs by applying the ML method using MEGA7 (Kumar et al. Citation2016) because one member contains only 11 PCGs, including five other species ().
Figure 1. Molecular phylogenetic analysis by Maximum Likelihood method. The evolutionary history was inferred using the Maximum Likelihood method based on the Tamura 3-parameter model. The tree with the highest log likelihood (-13987.63) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 6 nucleotide sequences. Codon positions included were first. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA7. Nucleotide sequence of the Geothelphusa sp. DJL-2014 isolate Gm01 (MG674171), G. dehaani (AB187570), Sinopotamon yangtsekiense (JF909980), and Sinopotamon xiushuiense (KU042041) were used. Eriocheir japonica (FJ455505) and Xenograpsus testudinatus (EU727203) were selected as outgroups.
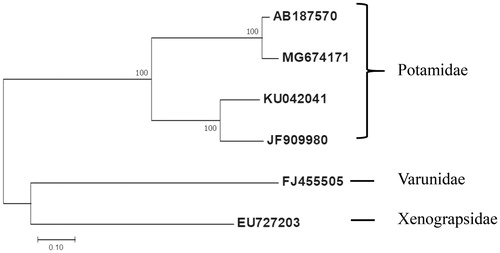
The Geothelphusa sp. DJL-2014 isolate Gm01 forms a clade with G. dehaani (AB187570). Sinopotamon yangtsekiense (JF909980) and Sinopotamon xiushuiense (KU042041) are the sister group of the Geothelphusa sp. DJL-2014 isolate Gm01. Eriocheir japonica (FJ455505) and Xenograpsus testudinatus (EU727203) were selected as outgroup. The atp8 gene is most divergent (92.3% identity) among 13 PCGs between Geothelphusa sp. DJL-2014 isolate Gm01 and G. dehaani ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogen Evol. 69:313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Lee DJ, Kim I-C. 2019. The new freshwater crab belongs to the genus Geothelphusa as inferred from mitochondrial and nuclear DNA markers. Indian J Anim Sci. Accepted recently.
- Liu Y, Cui Z. 2010. Complete mitochondrial genome of the Asian paddle crab Charybdis japonica (Crustacea: Decapoda: Portunidae): gene rearrangement of the marine brachyurans and phylogenetic considerations of the decapods. Mol Biol Rep. 37:2559–2569.
- Segawa RD, Aotsuka T. 2005. The mitochondrial genome of the Japanese freshwater crab, Geothelphusa dehaani (Crustacea: Brachyura): evidence for its evolution via gene duplication. Gene. 355:28–39.
- Yeo DC, Ng PK, Cumberlidge N, Magalhaes C, Daniels SR, Campos MR. 2008. Global diversity of crabs (Crustacea: Decapoda: Brachyura) in freshwater. Hydrobiologia. 595:275–286.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
