Abstract
Sparganium stoloniferum is a perennial aquatic or marsh herb. In this study, we assembled and characterized the complete chloroplast (cp) genome of S. stoloniferum for the first time. The circular genome is 161,759 bp in length and exhibits a typical quadripartite structure of the large single-copy (LSC, 88,957 bp) and small single-copy (SSC, 19,030 bp) regions, separated by a pair of inverted repeats (IRs, 26,886 bp). It contains 113 unique genes, including 79 protein-coding genes, 30 tRNA, and four rRNA genes. The phylogenetic analysis within order Poales demonstrated that Sparganium is sister to Typha, supporting inclusion of Sparganium and Typha in the single Typhaceae.
Sparganium L. is a marsh or aquatic perennial genus including approximately 14 species. Based on morphological characters, Sparganium had been placed in the monotypic Sparganiaceae (Cook and Nicholls Citation1986; Chen Citation1992) or included in Typhaceae together with Typha L. (Müller-Doblies Citation1970; Kubitzki Citation1998). Recent molecular phylogenetic studies indicated that Sparganium is sister to Typha and place the two genera in the single family, Typhaceae (Givnish et al. Citation2011, Citation2018; Sulman et al. Citation2013).
To date, only the complete chloroplast (cp) genome of Typha latifolia L. has been reported from Typhaceae with lack of cp genome data of Sparganium. Sparganium stoloniferum (Buch.-Ham. ex Graebn.) Buch.-Ham. ex Juz. is of economic importance. In China, it is a common traditional Chinese medicine plant which has been used for activating blood circulation, relieving pain, and treating cardiovascular disease (Tang et al. Citation2018). Here, we reported complete cp genome of S. stoloniferum for the first time and constructed phylogenetic tree for intention of confirming its position within order Poales.
The sample of S. stoloniferum was collected from Xingjianghe National Wetland Park, Anshun, Guizhou, China and specimens were deposited in Guizhou University Museum (Voucher: 106°08′42.58″, 26°16′47.17″, 1,279 m, Yang et al. 2018001, GACP). The total genomic DNA was extracted from fresh leaves using a modified CTAB method (Doyle and Doyle Citation1987) and sequenced based on the Illumina Hiseq 2500 platform (San Diego, CA). Approximately 5 GB of paired-end (150 bp) raw short sequence data were generated. The GetOrganelle (Jin et al. Citation2018) was used to de novo assemble the complete cp genome with Typha latifolia as the reference. Genome annotation was performed using DOGMA (Wyman et al. Citation2004, http://dogma.ccbb.utexas.edu/), coupled with manual correction for protein-coding region boundaries. Genome map was drawn using OGDraw program (Lohse et al. Citation2013), and the annotated genomic sequence of S. stoloniferum was submitted to GenBank under the accession number MK460210.
The chloroplast genome sequence of S. stoloniferum was 161,759 bp in length, composing of LSC region of 88,957 bp, SSC region of 19,030 bp, and two IR copies 26,886 bp. The overall GC content is 36.8% with the LSC, SSC, and IR regions being 34.8%, 30.6%, and 42.5%, respectively. A total of 113 unique genes were successfully annotated containing 79 protein-coding genes, 30 tRNA genes, and four rRNA genes. Among the genes identified, nine genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, and rps16) contained one intron and three genes (clpP, rps12, and ycf3) comprised of two introns. In addition, 20 genes have double copies, and ycf1 is pseudogene.
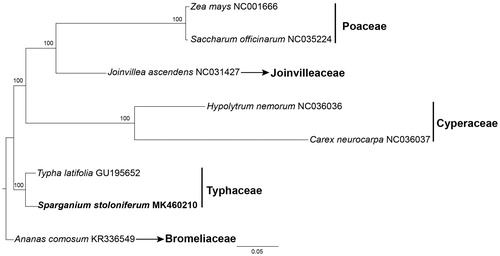
To validate the phylogenetic position of Sparganium within Poales, we made a phylogenomic analysis using RAxML-HPC2 on XSEDE v.8.1.11 (Stamatakis Citation2014) as implemented on the CIPRES computer cluster (http://www.phylo.org/) (Miller et al. Citation2010) with 1000 bootstrap replicates and Ananas comosus (L.) Merr. as the outgroup. The matrix contains seven completed cp genome representing five families of Poales. Phylogenomic analysis confirmed that Sparganium is sister to Typha with a strong support (100%), belonging to Typhaceae ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen YD. 1992. Sparganiaceae. In: Sun XZ, editor. Flora Reipublicae Popularis Sinicae. Vol. 8. Beijing: Science Press; p. 25–26.
- Cook CDK, Nicholls MS. 1986. A monographic study of the genus Sparganium (Sparganiaceae). Bot Helv. 96:213–267.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Givnish TJ, Barfuss MH, Van Ee B, Riina R, Schulte K, Horres R, Gonsiska PA, Jabaily RS, Crayn DM, Smith JAC, et al. 2011. Phylogeny, adaptive radiation, and historical biogeography in Bromeliaceae: insights from an eight-locus plastid phylogeny. Am J Bot. 98:872–895.
- Givnish TJ, Zuluaga A, Spalink D, Soto Gomez M, Lam VKY, Saarela JM, Sass C, Iles WJD, de Sousa DJL, Leebens-Mack J, et al. 2018. Monocot plastid phylogenomics, timeline, net rates of species diversification, the power of multi-gene analyses, and a functional model for the origin of monocots. Am J Bot. 105:1888–1910.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assemble of a complete circular chloroplast genome using genome skimming data. BioRxiv.
- Kubitzki K. 1998. Typhaceae. In: Kubitzki K, editor. Flowering plants. Monocotyledons: Alismatanae and Commelinanae (except Gramineae). Vol. 4. Heidelberg: Springer-Verlag; p. 457–461.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gate-way for inference of large phylogenetic trees. New Orleans, LA: Proceedings of the gateway computing environments workshop (GCE).
- Müller-Doblies D. 1970. Über die Verwandtschaft von Typha und Sparganium im Infloreszenz- und Blütenbau. Bot Jahrb. 89:451–562.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Sulman JD, Drew BT, Drummond C, Hayasaka E, Sytsma KJ. 2013. Systematics, biogeography, and character evolution of Sparganium (Typhaceae): diversification of a widespread, aquatic lineage. Am J Bot. 100:2023–2039.
- Tang YM, He C, Yang YJ, Duan JA, Wu QN, Liang QL. 2018. Research progress on ‘SanLeng’ from tubers of Scirpus yagara and Sparganium stoloniferum of different families. Modern Chinese Med. 20:110–116.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.