Abstract
Eothenomys, belongs to subfamily Arvicolinae, is a proper genus in China. The Yunnan red-backed vole, E. miletus, is an inherent species in Hengduan Mountain region. Here, the complete mitochondrial genome of E. miletus was sequenced (GenBank accession No. NC_030330). The complete mitochondrial sequence (16,344 bp) contains a non-coding region (D-loop) and 37 genes, including 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes. All PCGs are initiated by ATN codons, except for ND1 with GTG instead. Most of genes in E. miletus were distributed on the H-strand, except for the ND6 subunit gene and eight tRNA genes which were encoded on the L-strand. 13 PCGs of E. miletus were used to implement phylogenetic analyses with other 18 rodents.
It is subfamily Arvicolinae that have been regarded as ideal model for evolutionary studies of adaptation and speciation, as well as the role of Quaternary glacial cycles on diversification (Jaarola et al. Citation2004). Genus Eothenomys is predominantly distributed along the southeastern shoulder of the Qinghai-Tibetan Plateau, some of them are distributed in northeastern Burma, Assam and India (Luo et al. Citation2004; Liu et al. Citation2012). Due to the inherent morphological similarity and subjectivity regarding the descriptions of some species among this group, the taxonomy of the genus Eothenomys is under considerable debate. Although there are some genetic evidences to uncover the phylogeny of Eothenomys and discuss the independence of some species of this group (Luo et al. Citation2004; Liu et al. Citation2012), much more molecular markers are still needed to further research the evolution and phylogeny of this group. To date, partial sequence of the mtDNA of Eothenomys is available, only two complete mitochondrial sequences of this group, E. melanogaster and E. chinensis, were published (Yang et al. Citation2012; Chen et al. Citation2016). Considering the advantage of mitochondrial genome, for example, small size, maternal inheritance, accelerated rate of mutation compared to the nuclear DNA, as well as little or no recombination, it has been used as a star molecule for researching phylogenetic relationships at several taxonomic levels (Brown et al. Citation1979; Ballard and Whitlock Citation2004). We have sequenced the complete mitochondrial genome of E. miletus for contributing to establishing the database, which will be useful for resolving the taxonomy of genus Eothenomys in the future, and uncover the mechanism of speciation and adaptive evolution.
The E. miletus tissue specimen used in this study was collected from the field in Kunming (24°51'30''N 102°52'36''E), Yunnan Province, which was stored at College of Life Sciences, Yunnan Normal University. The genomic DNA was extracted from the liver tissue using a Tissue DNA Kit (BioTeke Corporation, Beijing, China). The complete genome sequence of E. miletus was sequenced using Sanger dideoxy sequencing. The complete mitochondrial sequence (16,344 bp) contains a non-coding region (D-loop) and 37 genes, including 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes. All PCGs are initiated by ATN codons, except for ND2 with GTG instead. Overall base composition of the complete mitochondrial DNA indicated a strong AT bias (Shadel and Clayton Citation1997), namely A (32.9%), T (26.2%), G (13.5%), C (27.5%); the percentage of A + T (59%) is higher than G + C (41%). Most mitochondrial genes are encoded on the H-strand except for ND6 and eight tRNA genes.
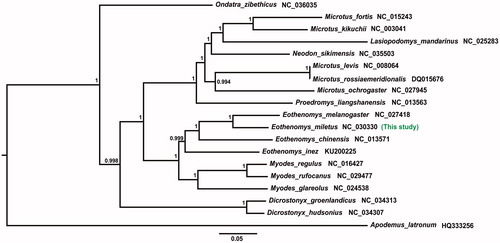
The phylogenetic relationship was reconstructed using Bayesian inference method (Mrbayes version 3.2.2) (Huelsenbeck and Ronquist Citation2001) based on 13 PCGs from 19 rodents. The best fitting model of sequence evolution was GTR + GAMMA, which was selected using MrModelTest version 2.3 according to Akaike Information Criterion (AIC) (Nylander et al. Citation2004). The phylogenetic tree in shows the relationship among 18 species of subfamily Arvicolinae, Apodemus latronum as the outgroup. Our result uncovers the significant support for Eothenomys had close relationship with Myodes, which was consistent with previous study (Lu et al. Citation2017). Besides, our result supported the Allen’s classification, in which E. inez is a species of subgenus Caryomys that belongs to genus Eothenomys (Allen Citation1940). Interestingly, the paraphyly relationship exists between genus Microtus and genus Lasiopodomys, Neodon. Our research provides some genetic backgrounds to further research the phylogeny and evolution of subfamily Arvicolinae.
Acknowledgements
We thank Jiahao Fu, Fangyan Ye and Zhuangqiong Ma for collecting the sample. We are especially grateful to Jiwei Qi and Xuelin Song for valuable suggestion. We sincerely thank the animals used in this research.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Allen GM. 1940. The mammals of China and Mongolia, Part II. Am Mus Nat Hist. 11:820–823.
- Ballard JWO, Whitlock MC. 2004. The incomplete natural history of mitochondria. Mol Ecol. 13:729–744.
- Brown WM, George M, Wilson AC. 1979. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci USA. 76:1967–1971.
- Chen X, Xiang D, Arunrugstichai S, Cai L, Xu Y. 2016. Complete mitochondrial genome of the Père David's Vole, Eothenomys melanogaster (Rodentia: Arvicolinae). Mitochondrial DNA. 27:1–2.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Jaarola M, Martínková N, Gündüz I, Brunhoff C, Zima J, Nadachowski A, Amori G, Bulatova NS, Chondropoulos B, Fraguedakis-Tsolis S, et al. 2004. Molecular phylogeny of the speciose vole genus Microtus (Arvicolinae, Rodentia) inferred from mitochondrial DNA sequences. Mol Phylogenet Evol. 33:647–663.
- Liu S, Liu Y, Guo P, Sun Z, Murphy RW, Fan Z, Fu J, Zhang Y. 2012. Phylogeny of Oriental voles (Rodentia: Muridae: Arvicolinae): molecular and morphological evidence. Zool Sci. 9:610–622.
- Lu T, Zhu M, Yi C, Si C, Yang C, Chen H. 2017. Complete mitochondrial genome of the gray red-backed vole (Myodes rufocanus) and a complete estimate of the phylogenetic relationships in Cricetidae. Mitochondrial DNA A. 28:62–64.
- Luo J, Yang D, Suzuki H, Wang Y, Chen W, Campbell KL, Zhang Y. 2004. Molecular phylogeny and biogeography of Oriental voles: genus Eothenomys (Muridae, Mammalia). Mol Phylogenet Evol. 33:349–362.
- Nylander JAA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL. 2004. Bayesian phylogenetic analysis of combined data. Syst Biol. 53:47–67.
- Shadel GS, Clayton DA. 1997. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 66:409–435.
- Yang C, Hao H, Liu S, Liu Y, Yue B, Zhang X. 2012. Complete mitochondrial genome of the Chinese oriental vole Eothenomys chinensis (Rodentia: Arvicolinae). Mitochondrial DNA. 23:131–133.