Abstract
The complete chloroplast genome sequence of Paeonia suffruticosa was determined by next-generation sequencing. The total length of the plastid genome of P. suffruticosa is 152,811 bp, including a large single-copy region (LSC) of 84,466 bp, a small single-copy region (SSC) of 17,051 bp, and a pair of identical inverted repeat regions (IRs) of 25,647 bp. A total of 111 genes was annotated, resulting in 77 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The phylogenetic analysis of P. suffruticosa with related chloroplast genome sequences in this study provided the close taxonomical relationship to Paeonia delavayi and Paeonia ludlowii in the genus Paeonia.
The genus Paeonia, the members of which are medicinal plants, is the only genus of the family Paeoniaceae and consists of around 30 species. Paeonia suffruticosa, the most important medicinal plant in this genus, has been distributed in China, Korea, and Japan, and has been cultivated for over 2000 years (Hong et al. Citation2001; Zhou et al. Citation2014; Zhao et al. Citation2016). The dried roots of this plant were used in traditional medicine in East Asia and have anti-aggregatory and blood anti-coagulant effects (Koo et al. Citation2010; Zhao et al. Citation2016), inhibition of nitric oxide production (Ding et al. Citation2012), and cellular apoptosis protection (Mukudai et al. Citation2013). To date, the genetic marker system to identify the species of Paeonia has not been well established (Hosoki et al. Citation1997; Wang JX et al. Citation2009; Wang S et al. Citation2014), and the genomic information has not been sufficient for reasonable position of this species among the species of Paeonia. We generated the complete chloroplast genome sequence of P. suffruticosa to provide a basic genetic resource and to help in authenticating species in the genus Paeonia.
The leaves of P. suffruticosa were collected from the Research Garden in Chungbuk National University (36° 37′ 28.61′′N, 127°27′ 18.36′′E), and the dried Plant specimen (NIBRVP0000634112) was deposited at the Herbarium in National Institute of Biological Resources (KB), Incheon, Korea. An Illumina paired-end (PE) genomic library with an average of 670 bp was constructed and sequenced using an Illumina HiSeq platform by Theragen (Suwon, Korea). A total of 26.7 Gb high-quality PE reads was obtained and assembled using a CLC Genomics Workbench version 7.5 (CLC Inc., Rarhus, Denmark). The genes in the chloroplast genome were annotated using the DOGMA programme (Wyman et al. Citation2004) and manual curation based on BLAST searches.
The complete plastid genome of P. suffruticosa (Genbank accession no. MH793271) has a circular molecular structure of 152,811 bp in length with 38.35% of GC content. Like typical chloroplast genomes, the complete chloroplast genome of P. suffruticosa was comprised of four regions: a large single-copy (LSC) region of 84,466 bp, a small single-copy (SSC) region of 17,051 bp, and a pair of identical inverted repeat (IR) regions (IRa and IRb) of 25,647 bp. We annotated 111 genes including 77 protein-coding genes, 30 tRNA genes, and 4 rRNA genes.
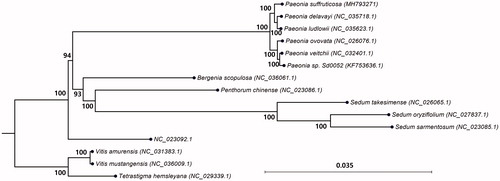
We performed phylogenetic analysis using the complete chloroplast genome sequence of P. suffruticosa with those of 14 species, which included 11 ingroup species of the Saxifragales, to which Paeonia belongs, and three outgroup species of the Vitales. For the tree generation, we used the time reversal model of the maximum likelihood algorithm with 1000 replications of bootstrap in CLC Genomics Workbench version 11.0 (CLC Qiagen). Six taxa of Paeoniaceae showed a monophyletic group, separated from six other species of Saxifragales. The phylogenetic tree placed P. suffruticosa close to Paeonia delavayi and Paeonia ludlowii and a relatively distant taxonomical relationship with Vitis species within the Vitaceae family ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ding L, Zhao F, Chen L, Jiang Z, Liu Y, Li Z, Qiu F, Yao X. 2012. New monoterpene glycosides from Paeonia suffruticosa Andrews and their inhibition on NO production in LPS-induced RAW 264.7 cells. Bioorg Med Chem Lett. 22:7243–7247.
- Hong DY, Pan KY, Turland NJ. 2001. Paeoniaceae. Flora Chin. 6:127–132.
- Hosoki T, Kimura D, Hasegawa R, Nagasako T, Nishimoto K, Ohta K, Sugiyama M, Haruki K. 1997. Comparative study of tree peony (Paeonia suffruticosa Andr.) cultivars and hybrids by random amplified polymorphic DNA (RAPD) analysis. J Jpn Soc Hortic Sci. 66:393–400.
- Koo YK, Kim JM, Koo JY, Kang SS, Bae K, Kim YS, Chung JH, Yun-Choi HS. 2010. Platelet anti-aggregatory and blood anti-coagulant effects of compounds isolated from Paeonia lactiflora and Paeonia suffruticosa. Pharmazie. 65:624–628.
- Mukudai Y, Kondo S, Shiogama S, Koyama T, Li C, Yazawa K, Shintani S. 2013. Root bark extracts of Juncus effusus and Paeonia suffruticosa protect salivary gland acinar cells from apoptotic cell death induced by cis-platinum (II) diammine dichloride. Oncol Rep. 30:2665–2671.
- Wang JX, Xia T, Zhang JM, Zhou SL. 2009. Isolation and characterization of fourteen microsatellites from a tree peony (Paeonia suffruticosa). Conserv Genet. 10:1029–1031.
- Wang S, Xue J, Ahmadi N, Holloway P, Zhu F, Ren X, Zhang X. 2014. Molecular characterization and expression patterns of PsSVP genes reveal distinct roles in flower bud abortion and flowering in tree peony (Paeonia suffruticosa). Can J Plant Sci. 94:1181–1193.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhao DD, Jiang LL, Li HY, Yan PF, Zhang YL. 2016. Chemical components and pharmacological activities of terpene natural products from the Genus Paeonia. Molecules. 21:1362.
- Zhou SL, Zou XH, Zhou ZQ, Liu J, Xu C, Yu J, Wang Q, Zhang DM, Wang XQ, Ge S, et al. 2014. Multiple species of wild tree peonies gave rise to the ‘king of flowers’, Paeonia suffruticosa Andrews. Proc Biol Sci. 281:20141687.