Abstract
Horornis fortipes has been listed on the IUCN red list of endangered species - Least Concern (LC). In this study, we sequenced the complete mitochondrial genome of H. fortipes by the polymerase chain reaction (PCR) method. The complete mitochondrial genome of H. fortipes is a circular molecule of 16,908 bp in total length, and it contains 13 typical vertebrate protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes (12srRNA, 16srRNA), and 2 control regions. This is the fist complete mitochondrial genome reported in Cettiidae. Phylogenetic analysis showed that Cettiidae diverged earlier than other family except Pycnonotidae, Acrocephalidae, and Pnoepygidae among 15 families within Passeriformes.
Horornis fortipes is a kind of resident bird which is distributed in southwestern areas and southeastern coastal areas of China, the Indian subcontinent, Indochina, and the Pacific islands (Zheng Citation2002). H. fortipes belongs to the genus Horornis within the family Cettiidae, and it has been listed on the IUCN red list of endangered species - Least Concern (LC). The Cettiidae is a family of Sylvioidea. Sylvioidea is one of the three superfamilies (Muscicapoidea, Sylvioidea, Passeroidea) recognized within the largest avian radiation, the Parvorder Passerida (Alström et al. Citation2006; Alström et al. Citation2013).
Mitochondrial DNA (mtDNA) has been widely used in molecular ecology, phylogeography study and many intractable phylogenies (Ballard and Whitlock Citation2004; Shen et al. Citation2010). In this study, we determined the complete mitochondrial genome of H. fortipes from an individual collected from the Laojun Moutain, Sichuan Province, China. The sample was kept in the Key Laboratory of Bioresources and Ecoenvironment, Sichuan University. The total DNA was extracted from the muscle tissue.
The complete mitochondrial genome sequence of H. fortipes is 16,908 bp in total length (GenBank Accession number MK051002). The overall composition of the base is 29.81% A, 23.82% T, 31.94% C, and 14.43% G, with an A + T ratio of 53.63%. And it contains 13 typical vertebrate protein-coding genes, 22 tRNA genes, 2 rRNA genes (12S rRNA, 16S rRNA), and 2 control regions, which is the same as some Sylvioidea birds (Qing et al. Citation2015; Jiao et al. Citation2018). The sizes of two control regions of H. fortipes mitochondrial genome are 963 bp (control regions 1) and 375 bp (control regions 2) respectively, with a longest identical sequence of 21 bp.
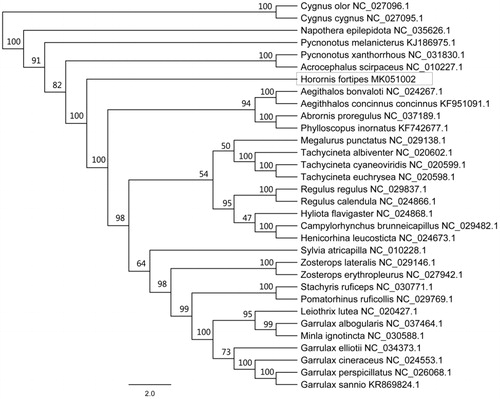
Figure 1. Phylogenetic tree of maximum likelihood (ML) method based on the nucleotide sequences of 13 mitochondrial PCGs of H. fortipes and 31 other species. Numbers represent node supports inferred from bootstrap support values.
The start codon of 13 protein-coding genes is ATG except for COI which is GTG. ND2, COII, ATP6, ATP8, ND3, ND4L, ND, and Cytb genes stop with TAA. The ND1, COI, and ND6 genes end with AGA, AGG, and TAG, respectively. And the stop codon of the COIII gene and ND4 gene are incomplete single-T. Among all genes, ND6 and eight tRNA genes (tRNA-Pro, tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Try, tRNA-Ser, and tRNA-Glu) are encoded on the L-strand, and others (12 protein-coding genes, 2 rRNAs, and 14 tRNAs) are encoded on the H-strand.
In order to understand the phylogenetic position of Cettiidae within Passeriformes, we constructed the phylogenetic tree (maximum likelihood) based on the concatenated DNA sequence of 13 mitochondrial PCGs of H. fortipes, 29 species from 14 other families of Passeriformes and 2 anseriformes species(outgroups). The result showed that Cettiidae diverged earlier than other families except for Pycnonotidae, Acrocephalidae, and Pnoepygidae among 15 families within Passeriformes. The phylogenetic positions of some families could not be solved due to low bootstrap support values. It was delusory that Garrulax albogularis did not cluster to Garrulax but cluster with Minla ignotincta ().
Acknowledgments
We would like to thank Mr. Guo Cai at Sichuan University for assistance in specimens collection in the field.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alström P, Ericson P, Olsson U, Sundberg P. 2006. Phylogeny and classification of the avian superfamily Sylvioidea. Mol Phylogenet Evol. 38:381–397.
- Alström P, Olsson U, Lei FM. 2013. A review of the recent advances in the systematics of the avian superfamily Sylvioidea. Chinese Birds. 4:99–131.
- Ballard JWO, Whitlock MC. 2004. The incomplete natural history of mitochondria. Mol Ecol. 13:729–744.
- Jiao SY, Liu ZX, Yu F, Yao JY, Li YM, Yan SQ. 2018. Complete mitochondrial genome of Pallas’s Leaf Warbler (Phylloscopus proregulus). Mitochondrial DNA Part B. 3:211–212.
- Qing H, Liu G, Zhou LZ, Wang JH, Li LY, Li B, Olsson U. 2015. Complete mitochondrial genome of Yellow-browed warbler Phylloscopus inornatus inornatus (Passeriformes: Sylviidae). Mitochondrial DNA. 26:939.
- Shen YY, Liang L, Sun YB, Yue BS, Yang XJ, Murphy R, Zhang YP. 2010. A mitogenomic perspective on the ancient, rapid radiation in the Galliformes with an emphasis on the Phasianidae. BMC Evol Biol. 10:132–141.
- Zheng GM. 2002. A cheklist on the classification and distribution of the birds of the world. 1st edition. Beijing, China: Science Press.
