Abstract
The complete chloroplast genome (cp) of Parnassia palustris was reported in this study. The cp genome was 148,700 bp in length including two inverted repeats (IRs) of 25,892 bp, which were separated by large single-copy (LSC) and small single-copy (SSC) of 82,175 and 14,741 bp, respectively. The GC content was 37%. The genome encoded 124 functional genes, including 78 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Using the whole chloroplast genome sequences alignment of 8 species from Celastraceae, Oxalidaceae, and Malpighiaceae, the phylogenetic relationship was built. The phylogenetic position of P. palustris was consistent with APG IV. The plastid genome of P. palustris will be useful for developing markers for further studies on resolving the relationship within the genus.
Parnassia L. (Linnaeus Citation1753) is a genus that is predominantly distributed in the arctic and temperate zones of the Northern Hemisphere; it is most speciose in China and the Himalayas (Simmons Citation2004). The genus was traditionally placed in the family Saxifragaceae (Hooker and Thompson Citation1857; Engler Citation1930; Cronquist Citation1981; Ku Citation1995; Ku and Hultgård Citation2001); however, it was recently transferred to Celastraceae (APG IV Citation2016; Ball Citation2016) based on molecular evidence (Soltis et al. Citation1997; Soltis et al. Citation2000; Zhang and Simmons Citation2006; APG IV Citation2016). Although as the type species of Parnassia, Parnassia palustris differs all the rest of genus Parnassia, having the multifid stamens and spreading widely in Europe, Asia, and North America.
In this study, fresh young leaves of P. palustris were collected from Yudushan, Beijing, China (N 40°32′45.55″, E 115°53′47.38″). The voucher specimens (YW-001) were deposited in the Herbarium of Beijing Forestry University (BJFC). Genomic DNAs were extracted using CTAB method (Doyle and Doyle Citation1987), and 2 × 150 bp pair end sequencing was performed on an Illumina HiSeq 4000 platform at Novogene (http://www.novogene.com, China). We used Map to Reference function in Geneious 11.1.5 (Kearse et al. Citation2012) to exclude nuclear and mitochondrial reads using published plastid genome of P. brevistyla (MG792145) as reference. Putative chloroplast reads were used for de novo assembling construction using Geneious version 11.1.5. Generated contigs were annotated and concatenated into larger ones based on the reference of P. brevistyla using Geneious version 11.1.5. The original data were again mapped to the larger contigs to extend their boundaries until all contigs were able to concatenate to one contig. The IR region was determined using the Repeat Finder function in Geneious version 11.1.5 and was inverted and copied to construct the complete chloroplast sequence. The chloroplast (cp) genome was manually adjusted to remove ambiguous sites. The annotation process was performed following by Liu et al. (Citation2018) using Morus mongolica (KM491711) as the reference. The plastome sequence of P. palustris was 148,700 bp in length containing a large single-copy (LSC) region of 82,175 bp, a small single-copy (SSC) region of 14,741 bp, and two inverted repeats (IR) of 25,892 bp. The genome contained 124 functional genes, including 78 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The sequence GC content was 37%.
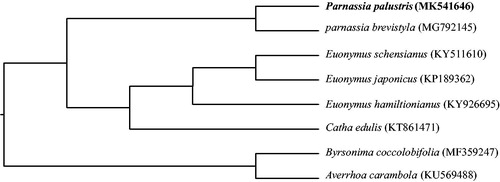
The Bayes inference phylogenetic tree was constructed based on the complete genomes of P. palustris, five species from Celastraceae, one species from Oxalidaceae and one species from Malpighiaceae (). The result was congruent with APG IV (Citation2016) and showed that P. palustris representing genus Parnassia belongs to Celastraceae with strong support.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- APG IV. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Ball PW, 2016. Parnassia. In: M. JS, Ball PW, Levin GA, editors. Flora of North America. Vol. 12. New York (NY): Oxford University Press; p. 113–117.
- Cronquist A. 1981. The integrated system of classification of flowering plants. New York (NY): Columbia University Press; p. 1–1262.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Engler A. 1930. Saxifragaceae. In: Engler A, Prantl K, editors. Die natürlichen pflanzenfamillien 18a. Leipzig, Germany: W Englmann; p. 178–182.
- Hooker JD, Thomson T. 1857. Praecursores ad floram Indicam. J Proc Linn Soc Bot. 2:54–103.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Ku TC, Hultgård UM. 2001. Parnassia. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 8. Beijing, China: Missouri Botanical Garden Press; p. 95–118.
- Ku TC. 1995. Parnassia. In: Lu LT, Huang SM, editors. Flora reipublicae popularis sinicae. Vol. 35. Beijing, China: Science Press; p. 1–66.
- Linnaeus C. 1753. Species plantarum. Stockholm, Sweden: Laurentius Salvius; p. 273.
- Liu H, He J, Ding C, Lyu R, Pei L, Cheng J, Xie L. 2018. Comparative analysis of complete chloroplast genomes of Anemoclema, Anemone, Pulsatilla, and Hepatica revealing structural variations among genera in tribe Anemoneae (Ranunculaceae). Front Plant Sci. 9:1097.
- Simmons MP. 2004. Parnassiaceae. In: Kubitzki K, editor. The families and genera of vascular plants. Berlin, Germany: Springer Verlag; p. 291–296.
- Soltis DE, Soltis PS, Chase MW, Mort ME, Albach DC, Zanis M, Savolainen V, Hahn WH, Hoot SB, Fay MF, et al. 2000. Angiosperm phylogeny inferred from a combined data set of 18S rDNA, rbcL, and atpB sequences. Bot J Linn Soc. 133:381–461.
- Soltis DE, Soltis PS, Nickrent DL, Johnson LA, Hahn WJ, Hoot SB, Sweere JA, Kuzoff RK, Kron KA, Chase MW, et al. 1997. Angiosperm phylogeny inferred from 18 S ribosomal DNA sequences. Ann Missouri Bot Gard. 84:1–49.
- Zhang LB, Simmons MP. 2006. Phylogeny and delimitation of the Celastrales inferred from nuclear and plastid genes. Syst Bot. 31:122–137.