Abstract
The complete mitochondrial genome of the lynx spider Oxyopes hupingensis was characterized in this study. The circular genome is 15,078 bp in length (GenBank accession number MK518391), containing 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA genes, and a control region. Its gene direction and arrangement are identical to those of other spider mitogenomes. The overall base composition is 34.76% A, 43.11% T, 8.04% C, and 14.09% G, showing AT-rich feature (77.87%). Ten PCGs are initiated with typical ATN start codons, three genes (cox2, cox3, and nad4) begin with TTG. Nine PCGs stop with complete termination codon TAA and TAG, cox1, nad4, nad4L, and cob are terminated with T. Twenty-three reading frame overlaps and four intergenic regions were found in the mitogenome of O. hupingensis. Three tRNAs lack the dihydrouracil (DHU) arm, whereas eight tRNAs lose the TΨC arm stems. Phylogenetic tree based on concatenated amino acid sequences of 13 PCGs shows that O. hupingensis has a close phylogenetic relationship with Oxyopes sertatus and belongs to Oxyopidae.
Keywords:
The lynx spider belongs to the family Oxyopidae, which is a small family among the known spiders and includes about 457 described species in 9 genera. Most of these species capture their prey directly and make little use of webs (Stávale et al. Citation2011). Many lynx spiders are important predators in agricultural systems as biological control agents (Meng et al. Citation2014). In this study, the samples of lynx spider Oxyopes hupingensis were collected from Libo county, Guizhou Province, China (E107°58′, N25°16′). The whole body specimen was preserved in ethanol and stored in the spider specimen room of Guiyang University with an accession number GYU-GZML-04.
The complete mitogenome of O. hupingensis (GenBank accession number MK518391) is 15,078 bp in length, containing 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rrnL and rrnS), and a putative control region. The gene direction and arrangement are identical to those observed in other spider mitogenomes (Pan et al. Citation2016; Xu et al. Citation2019). Fifteen mitochondrial genes were encoded on the minor strand (N-strand), while the rest of them were encoded on the major strand (J-strand). The overall base composition is 34.76% A, 43.11% T, 8.04% C, and 14.09% G, with A + T content of 77.87%. The AT-skew and GC-skew of this genome were −0.107 and 0.274, respectively.
Gene overlaps in O. hupingensis mitogenome were found in 23 locations and involved a total of 208 bp. The longest overlap is 31 bp in length and located between trnW and trnY. There are 4 intergenic spacer regions comprising a total of length of 18 bp and the largest spacer (6 bp) resided between trnP and nad6. The length of 22 tRNAs varied from 50 bp (trnA and trnR) to 66 bp (trnC), and A + T content varied from 66.00% (trnR) to 85.94% (trnD). Eleven tRNAs could not be folded into typical cloverleaf secondary structures. Eight of them (trnM, trnK, trnL2, trnR, trnF, trnP, trnT, and trnL1) lose the TΨC arm stem, three tRNAs (trnA, trnS1, and trnS2) lack the dihydrouracil (DHU) arm. The rrnL and rrnS were located between trnL1 and trnQ, and spaced by trnV, with a length of 1008 and 697 bp, respectively. The control region was located between trnQ and trnM with 1500 bp length and 77.27% A + T content.
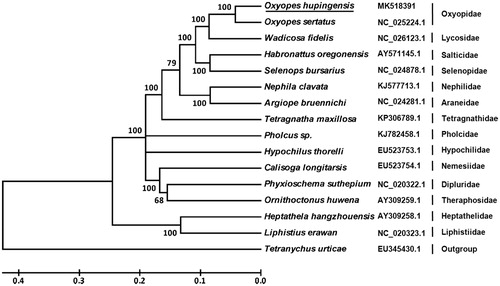
Ten PCGs are initiated with typical ATN start codons (ATA and ATT), three genes (cox2, cox3, and nad4) start with TTG. Nine PCGs end with two types of complete termination codon, TAA (cox2, cox3, atp6, atp8, nad2, nad3, nad5, and nad6) and TAG (nad1). The remaining PCGs end with incomplete termination codon (T), including cox1, nad4, nad4L, and cob. Based on the concatenated amino acid sequences of 13 PCGs, the neighbor-joining method was used to construct phylogenetic relationship of O. hupingensis and 14 other spiders. The results showed that O. hupingensis is closely related to Oxyopes sertatus (), which agree with the morphology taxonomy of the family Oxyopidae.
Disclosure statement
The authors report no conflicts of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Meng X, Ouyang GC, Liu H, Hou BH, Huang SS, Guo MF. 2014. Molecular screening and predation evaluation of the key predators of Conopomorpha sinensis Bradley (Lepidoptera: Gracilariidae) in litchi orchards. Bull Entomol Res. 104:243–250.
- Pan WJ, Fang HY, Zhang P, Pan HC. 2016. The complete mitochondrial genome of striped lynx spider Oxyopes sertatus (Araneae: Oxyopidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1616–1617.
- Stávale LM, Schneider MC, Brescovit AD, Cella DM. 2011. Chromosomal characteristics and karyotype evolution of Oxyopidae spiders (Araneae, Entelegynae). Genet Mol Res. 10:752–763.
- Xu KK, Lin XC, Yang DX, Yang WJ, Li C. 2019. Characterization of the complete mitochondrial genome sequence of Neoscona scylla and phylogenetic analysis. Mitochondrial DNA Part B. 4:416–417.