Abstract
Hickory (Carya cathayensis Sarg.) is an important economic plant and native to Anhui and Zhejiang Province, China. The complete chloroplast (cp) genome sequence of C. cathayensis was determined by Next-generation sequencing technology. The total length of the cp genome is 160,666 bp and contains 86 protein-coding genes, 39 tRNA genes, and 8 rRNA genes. Phylogenetic analysis suggested that Carya is the sister group of the genus Annamocarya within the walnut family. The DNA data presented here will be useful to study the genetic diversity and conservation genetics of C. cathayensis.
Hickory (Carya cathayensis Sarg.) is a member of the walnut family, Juglandaceae and native to eastern China, mainly distributed near Tianmu Mountain, south of Anhui Province and northeast of Zhejiang Province (Wang and Fang Citation1991). It is a commercially valuable tree for the edible nuts, which has a long juvenile phase. This characteristic is recognized as a disadvantage in breeding and in stable annual production (Wang et al. Citation2012). Now, most of the plants are cultivated and some of them are being hybridized with other individuals to improve the production. Some molecular data of the natural population were determined (Guo et al. Citation2007; Wang et al. Citation2012; Xiang et al. Citation2014). The study of the cp genome of the wild-type C. cathayensis was determined and described in order to provide basic genetic information about this species.
The fresh leaves (Voucher number: HSP2018001) of a juvenile C. cathayensis were sampled from the campus of Huangshan University. The wild juvenile plant was transplanted from Shexian County, Anhui Province, China. The cp genome sequence of C. cathayensis was determined by Next-generation sequencing technology (Illumina Hiseq 2500, Shanghai Majorbio Bio-pharm Technology Co., Ltd. (SMBTC, Shanghai, China)). Genome sequences were picked out and assembled by the software SOAPdenovo v2.04 (Luo et al. Citation2012). The cp genome of C. cathayensis (Genbank accession number MK414769) showed a typical structure with a length of 160,666 bp, which contained a large single-copy (LSC) and a small single copy (SSC) of 90,025 bp and 20,979 bp, respectively, separated by the inverted repeats (IR) of 24,831 bp. The cpDNA is structured with 130 genes, comprising 86 protein-coding genes, 39 tRNA genes, and 8 rRNA genes. The overall base composition of the entire genome is as follows: A (31.5%), T (32.3%), C (18.4%) and G (17.8%), which the percentage of A + T is 63.8%.
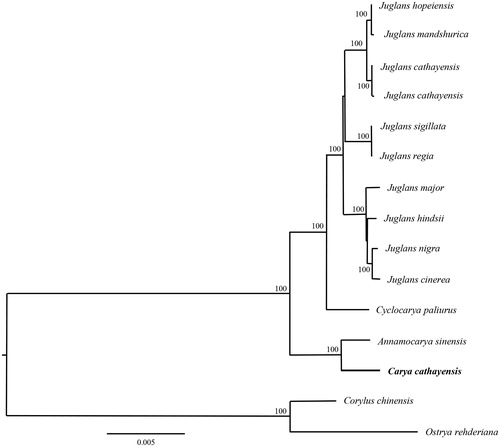
In order to validate the newly determined sequence, the whole chloroplast genome sequence of the C. cathayensis and together with other 13 closely related species sequences from GeneBank to perform phylogenetic analysis. A maximum likelihood (ML) tree was constructed based on the dataset by the online tool RAxML (Kozlov et al. Citation2018). Phylogenetic analysis result suggested that Carya is the sister group of the genus Annamocarya within the walnut family () and was consistent with the previous research (Xiang et al. Citation2014).
Figure 1. The phylogenetic tree (ML) was constructed based on the dataset of whole chloroplast genome. The numbers above the branch meant bootstrap value. The analyzed species and corresponding NCBI accession number as follows: Juglans hopeiensis (KX671977), Juglans mandshurica (MF167461), Juglans cathayensis (KX671976), Juglans cathayensis (MF167457), Juglans sigillata (KX424843), Juglans regia (KT870116), Juglans major (MF167460), Juglans hindsii (MF167459), Juglans nigra (MF167462), Juglans cinerea (MF167458), Cyclocarya paliurus (KY246947), Annamocarya sinensis (KX703001), Carya cathayensis (MK414769) Corylus chinensis (KX814336), Ostrya rehderiana (KT454094).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Guo C, Huang J, Wang Z, Fang Y. 2007. RADA analysis on the genetic diversity of Carya cathayensis germplasm resources. Acta Laser Biol Sinica. 16:405–410.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2018. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. BioRxiv 447110; doi:10.1101/447110
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 1:18.
- Wang S, Fang X. 1991. A brief account of walnut production in Anhui Province. Nonwood Forest Res. 9:33–37.
- Wang ZJ, Huang JQ, Huang YJ, Li Z, Zheng BS. 2012. Discovery and profiling of novel and conserved micrornas during flower development in Carya cathayensis via deep sequencing. Planta. 236:613–621.
- Xiang X-G, Wang W, Li R-Q, Lin L, Liu Y, Zhou Z-K, Li Z-Y, Chen Z-D. 2014. Large-scale phylogenetic analyses reveal fagalean diversification promoted by the interplay of diaspores and environments in the paleogene. Perspect Plant Ecol. 16:101–110.
