Abstract
Opisthotropis latouchii is an endemic species which inhabits flowing streams in China. In this study, the complete mitochondrial genome of O. latouchii was determined using high-throughput sequencing data. The mitogenome was 17,051 bp in length and contained 13 protein-coding genes, 22 transfer RNA, 2 ribosomal RNA and 2 control regions. The total nucleotide composition was 34.09% A, 25.35% T, 27.39% C, and 13.16% T, respectively. The phylogenetic analysis showed that O. latouchii was closely related with Nerodia sipedon in Colubridae.
Opisthotropis latouchii is an aquatic specialist snake that is distributed only in China. Little is known about this species although it has been considered as a national protected terrestrial wild animal with good benefits or important economic and scientific values since 2000. In this study, the complete mitochondrial genome of O. latouchii was sequenced and annotated.
The specimen of O. latouchii was collected from Gutian Mountain (Zhejiang province, China) and now is stored in the Zoological Laboratory of Zhejiang Normal University. Genomic DNA was extracted from muscle tissue using Rapid Animal Genomic DNA Isolation Kit (Sangon Biotech.). Paired-end sequencing was performed by MajorBio (Shanghai, China) on a Illumina HiSeqTM DNA sequencing platform. The mitogenome was assembled with MITObim (Hahn et al. Citation2013) using O. latouchii mitochondrial CYTB gene (GenBank: GQ281783) and ND4 gene (GenBank: JQ687421) as the starting seed, and the complete mitogenome of Nerodia sipedon (GenBank: JF964960) as the reference. And it was annotated by MITOS2 web server (Bernt et al. Citation2013).
The complete mitochondrial genome of O. latouchii is 17,051 bp in length (GenBank Accession No. MK570292) and the total nucleotide composition is 34.09% A, 25.35% T, 27.39% C, 13.16% T, with a slight AT bias of 59.44%. It contains 37 typical mitochondrial genes, including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA), two ribosomal RNA (12S rRNA and 16S rRNA) and two putative control regions (CRs). Among these genes, 28 of 37 genes are encoded on the H-strand, including 12 PCGs, 14 tRNA, and 2 rRNA. Nine other genes, ND6, tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAGlu, and tRNAPro, are encoded on the light strand. For the 13 PCGs, the longest gene is ND5 (1782 bp), while the shortest gene is ATP8 (165 bp). The start codons in PCGs are ATG (COXII, ATP8, ATP6, COXIII, ND4L, ND4, ND5, ND6, CYTB), ATT (ND2, ND3), ATA (ND1) or GTG (COXI). Moreover, seven PCGs end with stop codons TAA, TAG or AGG, while six other PCGs use incomplete stop codon T. Additionally, two rRNA (925 bp and 1463 bp in length, respectively) are located between tRNAPhe and ND1 genes and are separated by tRNAVal. The two CRs are located between tRNAPro and tRNAPhe and between tRNAIle and tRNALeu(UUR), respectively. And they nearly have the same nucleotide composition except for 10 bp shorter than CR1 (972 bp) and 6 bp differences.
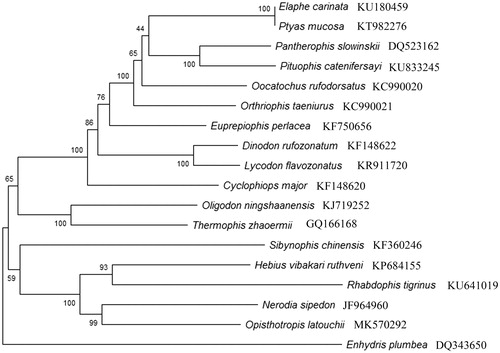
The phylogenetic relationship under Colubridae is recovered based on the concatenated of 13 PCGs from 17 other natricine snakes with Maximum Likelihood (ML) method using MEGA X software (Kumar et al. Citation2018). The results showed O. latouchii was closely related with Nerodia sipedon () and the relationships among 18 genera in Colubridae were showed similar topology as previous researches (Sun et al. Citation2017; Yang et al. Citation2019; Zhu et al. Citation2019).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Sun H, Li E, Sun L, Yan P, Xue H, Zhang F, Wu X. 2017. The complete mitochondrial genome of the greater green snake Cyclophiops major (Reptilia, Serpentes, Colubridae). Mitochondrial DNA Part B Resources. 2:309–310.
- Yang S, Shi Q, Tang W, Zhang C, Cheng Y, Zhu G. 2019. The complete mitochondrial genome sequence and phylogenetic analysis for Rhabdophis himalayanus (Squamata:Colubridae). Mitochondrial DNA Part B Resources. 4:705–706.
- Zhu G, Xu H, Tang W, Yang S, Shi Q, Cheng Y, Wang Q, He T. 2019. The complete mitochondrial genome and phylogenetic analysis of Rhabdophis adleri (Squamata: Colubridae). Mitochondrial DNA Part B Resources. 4:423–425.