Abstract
The complete mitochondrial genome (mitogenome) of the leafhopper species Paralaevicephalus gracilipenis (Dai et al. Citation2005) (Hemiptera: Cicadellidae: Deltocephalinae) are sequenced and annotated. The mitochondrial genome is 16,114 bp, and nucleotide composition of the whole mitogenome is highly A + T biased (A: 42.0%; T: 33.4%, G: 10.3%, C: 14.3). All PCGs started with ATN and stopped with TAN, except COX2, which started with TTG and stopped with single T. The phylogenetic tree confirms that P. gracilipenis and Yanocephalus yanonis are clustered into a clade. This study enriches the mitogenomes of the leafhopper subfamily Deltocephalinae.
The leafhopper genus Paralaevicephalus was established by Ishihara (Citation1953), it belongs to the tribe Paralimini of subfamily Deltocephalinae (Hemiptera: Cicadellidae). This genus now contains 12 species, all species are very similar in coloration and difficult to distinguish externally, but the structure of male genitalia are markedly different (Xing et al. Citation2009; Li et al. Citation2011). Moreover, all of them are closely associated with grass-dominated habitats. In this study, the complete mitochondrial genome sequences of Paralaevicephalus gracilipenis (Dai et al. Citation2005) were determined using next-generation sequencing method for the first time. This study will facilitate future studies on the identification, population genetics, and evolution of the leafhopper subfamily Deltocephalinae.
Total genome DNA was extracted from a male adult of P. gracilipenis which was collected from grass, in Wangcao, Suiyang County, Guizhou Province, China, in August 2016. And voucher specimen’s genome DNA and male external genitalia are deposited in the Institute of Entomology, Guizhou University, Guiyang, China (GUGC). The complete mitochondrial genome (mitogenome) of P. gracilipenis is 16,114 bp long (GenBank accession number MK450366), and contained the typical set of 37 typical mitochondrial genes (13 protein-coding genes, 22 tRNA genes, and 2 rRNA genes), and an A + T rich region. The nucleotide composition of the whole mitogenome is highly A + T biased (A: 42.0%; T: 33.4%, G: 10.3%, C: 14.3). All PCGs started with the canonical putative start codon ATN except for the ND5 gene which started with TTG instead, similar to other leafhoppers (Du et al. Citation2017). 12 genes shared complete termination codon TAA/TAG, while COX2 used incomplete stop codon (a single T), is a common phenomenon in Previous reports of the sequenced Deltocephalinae mitogenomes (Song et al. Citation2017).
All tRNA genes are identified by ARWEN version 1.2 software (Laslett and Canbäck, Citation2008). The 16S rRNA gene is 1232 bp long and located between tRNA-L2 and tRNA-V; the 12S rRNA gene is 758 bp length and located after tRNA-V. The control region is 1711 bp long and is located between 12S rRNA and tRNA-I, and there is a 792 bp repeat sequence within A + T rich region (264 bp repeat unit repeats thrice).
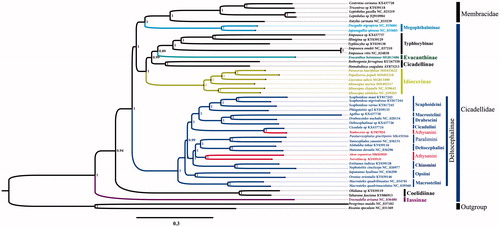
The phylogenetic relationships of P. gracilipenis were reconstructed using MrBayes analysis based upon the concatenated nucleotide sequences of the 13 PCGs and 2 rRNA genes (). Each PCG and rRNA sequences were aligned using MAFFT algorithm in the Translator X (http://pc16141.mncn.csic.es/index_v4.html) and MAFFT v7.0 (https://mafft.cbrc.jp/alignment/server/) with G-INS-i strategy respectively (Katoh et al. Citation2017), and poorly aligned site removed by Gblocks 9.1b (http://www.phylogeny.fr/one_task.cgi?task_type=gblocks) (Abascal et al. Citation2010; Castresana Citation2000). The dataset was used in the phylogenetic analysis by MrBayes software ((Miller et al., Citation2010) under the GTR + I + G model. The phylogenetic tree confirms that P. gracilipenis is part of the Paralimini, which clustered with Yanocephalus yanonis into a clade with strong support (posterior probability = 1) ().
Figure 1. Phylogenetic analyses of Paralaevicephalus gracilipenis based upon the concatenated nucleotide sequences of the 13 PCGs and 2 rRNAs of 47 species. The analysis was performed using MrBayes software. Numbers at nodes are bootstrap values. The accession number for each species is indicated after the scientific name.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucl Acids Res. 38:W7–W13.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Dai W, Zhang YL, Hu J. 2005. Review of the Chinese leafhopper genus Paralaevicephalus (Hemiptera: Cicadellidae: Deltocephalinae) with descriptions of two new species from China. Can Entomol. 137:404–409.
- Du Y, Dai W, Dietrich CH. 2017. Mitochondrial genomic variation and phylogenetic relationships of three groups in the genus Scaphoideus (Hemiptera: Cicadellidae: Deltocephalinae). Sci Rep. 7:16908.
- Ishihara T. 1953. A tentative check list of the superfamily Cicadelloidea of Japan (Homoptera). Sci Rep Matsuyama Agric Coll. 11:1–72.
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 1–7.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Li ZZ, Dai RH, Xing JC. 2011. Deltocephalinae from China (Hemiptera: Cicadellidae). Beijing: Popular Science Press; p. 336. [in Chinese with English summary].
- Miller, MA, Pfeiffer, W, Schwartz, T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedingsof the Gateway Computing Environments Workship (GCE). New Orleans. 1–8. DOI:10.1109/GCE.2010.5676129
- Song N, Cai WZ, Li H. 2017. Deep-level phylogeny of Cicadomorpha inferred from mitochondrial genomes sequenced by NGS. Sci Rep. 7:10429.
- Xing JC, Dai H, Li ZZ. 2009. A taxonomic study of the genus Paralaevicephalus Ishihara (Hemiptera: Cicadellidae: Deltocephalinae), with description of four new species from China. Zootaxa. 1979:53–61.
