Abstract
We sequenced and annotated the nearly complete mitochondrial genome (mitogenome) of Lemnia saucia (Coleoptera: Coccinellidae). This mitogenome was 14,106 bp long and encoded 13 protein-coding genes (PCGs), 19 transfer RNA genes, and two ribosomal RNA genes. The overall nucleotide composition was 41.6% of A, 8.8% of G, 37.3% of T, and 12.4% of C. Phylogenetic reconstruction using both Bayesian inference (BI) and maximum likelihood (ML) validated the taxonomic status of L. saucia, exhibiting the close relationship with Propylea japonica.
Lemnia saucia (Coleoptera: Coccinellidae) is a predatory ladybird, which is widely distributed in China. In this study, we sequenced most part of the mitochondrial genome of the L. saucia (GenBank accession No. MK574678), representing the first mitochondrial genome from the genus Lemnia.
Adult specimens of L. saucia were collected in Xixi national wetland park, China. The collected specimens were stored in 95% ethanol at temperature –20 °C and deposited in the College of Plant Protection, Nanjing Agricultural University, Nanjing, China. Whole genomic DNA was extracted from abdomen of each specimen using a Wizard® Genomic DNA Purification Kit (Promega, Madison, WI) according to the manufacturer’s instructions. The mitogenome of Propylea japonica (GenBank accession No. KM244660.1) was employed as the reference sequence (Tang et al. Citation2014). Certain pairs of universal primers for ladybird mitochondrial genomes were used for polymerase chain reaction (PCR) amplification (Simon et al. Citation2006). Then PCR products were sequenced using primer-walking strategy from both strands by Genscript Biotech Corp. (Nanjing, China). Mitochondrial genome was assembled by SeqMan program from DNASTAR (Burland Citation2000) and annotated using MITOS Web Server (Bernt et al. Citation2013).
We obtained partial mitogenome of L. saucia with 14,106 bp long. The region that we failed to sequence was between rrnS and nad2, and generally contained three transfer RNA genes (tRNAs; trnI, trnQ, and trnM) and a putative control region. This mitogenome encoded 13 protein-coding genes (PCGs), 19 tRNAs, two ribosomal RNA unit genes (rrnL and rrnS). The overall nucleotide composition was 41.6% of A, 8.8% of G, 37.3% of T, and 12.4% of C. Twelve PCGs started with typical ATN codon (two with ATA, four with ATG, and six with ATT), whereas the cox1 gene appeared to start with TTA. Eight PCGs terminated with TAN codon (seven with TAA and one with TAG), and the remaining five terminated with an incomplete stop codon TA or T.
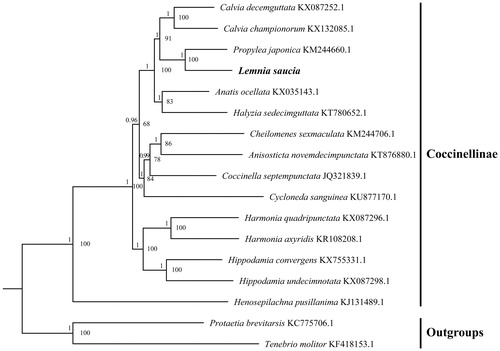
To validate the phylogenetic position of L. saucia, the ML and BI trees were constructed on CIPRES Portal using 13 mitochondrial PCGs from mitogenomes of 15 species and two outgroups from Scarabaeidae and Tenebrionidae, respectively. We used the best-fit partitioning scheme and partition-specific models recommended by PartitionFinder (Lanfear et al. Citation2012). Two phylogenetic analyses using different methods yielded the same topology, and nodal supporting values were always higher for BI trees than for ML trees. As shown in , L. saucia was positioned near P. japonica within the subfamily of Coccinellinae, which was in accordance with the previous molecular works (Aruggoda et al. Citation2010).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Aruggoda AG, Shunxiang R, Baoli Q. 2010. Molecular phylogeny of ladybird beetles (Coccinellidae: Coleoptera) inferred from mitochondrial 16S rDNA sequences. Trop Agric Res. 21:209–237.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Burland TG. 2000. DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol. 132:71–91.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
- Simon C, Buckley T, Frati F, Stewart J, Beckenbach A. 2006. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Syst. 37:545–579.
- Tang M, Tan M, Meng G, Yang S, Su X, Liu S, Song W, Li Y, Wu Q, Zhang A, et al. 2014. Multiplex sequencing of pooled mitochondrial genomes-a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42:e166.