Abstract
The complete mitogenome of Cynoglossus roulei is 16,598 bp in length, containing 37 genes, among them, ND6 and eight tRNA genes are encoded by L-strand and other genes by H-strand, which are as same as those of typical mitogenome in fishes. The gene rearrangement related to one tRNA gene and CR were found, forming the gene order of CR-Gln-Ile-Met, which is the same as that of mitogenomes in all identified Cynoglossus speices. Phylogenetic tree based on 12 protein coding genes, tRNA and rRNA shows that C. roulei has a closer phylogenetic relationship to C. semilaevis.
Cynoglossus roulei (Wu, 1932) belongs to Cynoglossinae of the family Cynoglossidae in the Pleuronectiformes, this species is distributed offshore of Hainan, Guangxi and Guangdong to the south of Zhoushan Islands of China (Li and Wang Citation1995). Because the morphological differences between Cynoglossidae fishes are not obvious, it is necessary to develop molecular markers to figure out controversial issues of their phylogenetic relationships (Miao et al. 2013). Analyzing the mitochondrial genome of C. roulei is a good case to find the possible molecular markers for the further researches.
The mitochondrial genome of C. roulei was obtained (GenBank accession number: MK574671) based on one sample collected from Taizhou City, Zhejiang, China (N 28°21′36.97″, E 121°37′10.04″). This specimen was stored in the Marine Biodiversity Collection of South China Sea, Chinese Academy of Sciences, under the voucher number SCF201510416.
The mitochondrial genome of C. roulei is 16,598 bp in length, including 13 protein-coding genes, two ribosomal RNA (rRNA), 22 transfer RNA (tRNA), and a control region (CR). Among these genes, 28 genes are encoded on the H-strand, others including ND6 and eight tRNA genes on the L-strand. The overall base composition is 31.26%, 24.95%, 14.25%, and 29.54% for A, C, G, T, respectively, with a bias on AT content (60.8%).
The size of 13 protein-coding genes ranges from 165 bp (ATP8) to 1854 bp (ND5), that of 12S and 16S rDNA are 946 bp and 1683 bp respectively, and that of CR is 853 bp. The size of 22 tRNA genes ranges from 66 bp (tRNA-Cys) to 75 bp (tRNA-Lys), which were identified using tRNAscan-SE 2.0 (Lowe and Chan Citation2016 (Miao, Citation2013)).
Additionally, a gene rearrangement related to one gene and CR was found in this study, which was similar to that of other Cynoglossus species (Kong et al. Citation2009; Shi, Jiang, et al. Citation2015; Wei et al. Citation2016), but different with that of Paraplagusia species (Shi, Gong, et al. Citation2015) In the mitogenome of C. roulei, CR translocated from the position between tRNA-Pro and tRNA-Phe to the position after ND1, and the tRNA-Gln translocated from the position between tRNA-Ile and tRNA-Met to the position before tRNA-Ile, which formed the genes order of CR-Gln-Ile-Met.
In this mitogenome, 15 intergenic spaces exist, with a total length of 148 bp, among them, the longest is 75 bp between tRNA-Gln and tRNA-Ile.
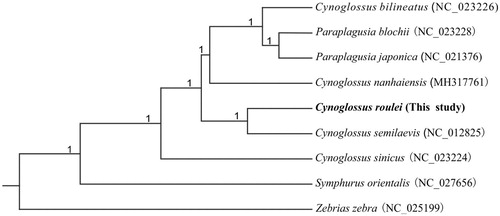
The phylogenetic tree was constructed based on the mitogenomes of C. roulei and other seven Cynoglossidae species with Zebrias zebra as outgroup by using Bayesian inference in MrBayes 3.2 (Ronquist and Huelsenbeck Citation2003). The mitogenome sequences of seven species and one outgroup were retrieved from GenBank. In the phylogenetic tree (), all species of Cynoglossinae formed one clade with 100% posterior probability. Cynoglossus roulei and Cynoglossus semilaevis formed a clade, which means C. roulei has a relatively closer phylogenetic relationship to C. semilaevis, comparing with other three Cynoglossus species.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Kong XY, Dong XL, Zhang YC, Shi W, Wang ZM, Yu ZN. 2009. A novel rearrangement in the mitochondrial genome of tongue sole, Cynoglossus semilaevis: control region translocation and a tRNA gene inversion. Genome. 52:975–984.
- Li SZ, Wang HM. 1995. Fauna sinica (Osteichthyes): pleuronectiformes. China. Beijing: Science Press.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Miao, XG, Jiang, JX, Shi, W, Wang, ZM, Wang, SY, KongXY. 2013. Phylogenetic relationship of Cynoglossinae based on COI barcodingmarker. Journal of Tropical Oceanography. 32:85–92.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Shi W, Gong L, Wang SY, et al. 2015. Tandem duplication and random loss for mitogenome rearrangement in Symphurus (Teleost: Pleuronectiformes). Bmc Genomics. 16:355.
- Shi W, Jiang JX, Miao XG, Kong XY. 2015. The complete mitochondrial genome sequence of Cynoglossus sinicus (Pleuronectiformes: Cynoglossidae). Mitochondrial DNA. 26:865–866.
- Wei M, Liu Y, Guo H, Zhao F, Chen S. 2016. Characterization of the complete mitochondrial genome of Cynoglossus gracilis and a comparative analysis with other Cynoglossinae fishes. Gene. 591:369–375.