Abstract
Suaeda japonica Makino is an annual herb in seashore in Korea and Japan. In this study, we presented the first complete chloroplast genome of S. japonica which is 152,109 bp long and has four subregions: 83,618 bp of large single copy (LSC) and 18,101 bp of small single copy (SSC) regions are separated by 25,195 bp of inverted repeat (IR) regions including 128 genes (83 protein-coding genes, eight rRNAs, and 37 tRNAs). The overall GC content of the chloroplast genome is 36.4% and those in the LSC, SSC, and IR regions are 34.2%, 29.2%, and 42.7%, respectively. Phylogenetic trees show that two Suaeda chloroplast genomes were clustered and clade of Betoidae, Chenopodioidae, and Amarantheae are successfully resolved with high bootstrap supports. Our chloroplast genome will contribute for phylogeny of Amaranthaceae in detail.
Around 100 species in genus Suaeda Forsskål ex J. F. Gmelin (Amaranthaceae), usually distributed in the seashore, wasteland, and around the lake, are annual, subshrubs, or shrubs in Europe, Asia, and North America. It has alternate, blade terete, semiterete or somewhat compressed, entire, succulent leaves (Chung Citation1992). There are five Suaeda species: S. glauca, S. japonica, S. australis, S. maritima, and S. malacosperma in Korea (Chung Citation2018).
Suaeda japonica Makino, an herb species, is growing up to 20 ∼ 50 cm with a woody stem. The stem is branched into 3 or 4 times from the middle part. Branches are ascending, long, and slender. It has fleshy leaves whose length ranges from 5 ∼ 35 mm and width is 2 ∼ 4 mm, which are green in young stage but turns into purple-red color later (Chung Citation1992). It is usually found in a tidal flat in the West Sea in South Korea and Kyushu in Japan (Hara and Kurosawa Citation1963; Chung Citation1992). We completed the whole chloroplast genome of S. japonica for unraveling its phylogenetic position.
Total DNA of S. japonica collected in Julpo, Buan-gun, Jeollabuk-do, Korea (Voucher in InfoBoss Cyber Herbarium (IN); IB-00598) was extracted from fresh leaves by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeqX at Macrogen Inc., Seoul, Korea. de novo assembly and confirmation were done by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.1.5 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation based on Suaeda malacosperma chloroplast complete genome (NC_039180; Park et al. Citation2018).
The chloroplast genome of S. japonica (Genbank accession is MK558824) is 152,109 bp (GC ratio is 36.4%) and has four subregions: 83,618 bp of large single copy (LSC; 34.2%) and 18,101 bp of small single copy (SSC; 29.2%) regions are separated by 25,195 bp of inverted repeat (IR; 42.7%). It contains 128 genes (83 protein-coding genes, eight rRNAs, and 37 tRNAs); 17 genes (six protein-coding genes, four rRNAs, and seven tRNAs) are duplicated in IR regions.
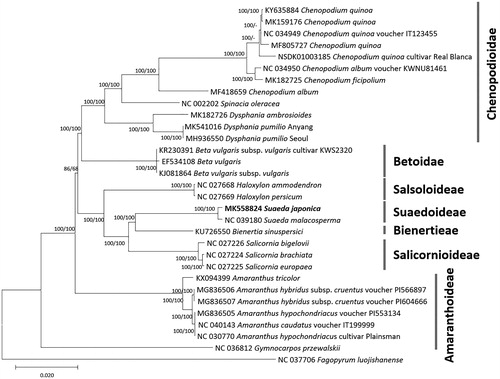
Thirty-one complete chloroplast genome sequences of Amaranthaceae including two Sueada chloroplast genomes (Kim et al. Citation2016; Devi and Thongam Citation2017; Hong et al. Citation2017; Park et al. Citation2018; Hong et al. Citation2019; Kim et al. Citation2019a, Citation2019b; Maughan et al. Citation2019) were aligned by MAFFT 7.388 (Katoh and Standley Citation2013) after rearranging SSC and LSC regions of Chenopodium quinoa ‘Real Blanca’ for constructing neighbor joining (bootstrap repeat is 10,000) and maximum likelihood trees (bootstrap repeat is 1,000) using MEGA X (Kumar et al. Citation2018). Phylogenetic trees show that two Suaeda chloroplast genomes were clustered with high bootstrap support (). In addition, three Beta chloroplast genomes in Betoidae are clustered with Chenopodioidae clade with high bootstrap values, improving unresolved clade of Betoidae, Chenopodioidae, and Amarantheae in previous phylogeny studies based on limited molecular markers (Kadereit et al. Citation2003; Kapralov et al. Citation2006). Altogether, our chloroplast genome will contribute to understanding the phylogeny of Amaranthaceae in detail.
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of 31 Amaranthaceae complete chloroplast genomes: Suaeda japonica (MK558824 in this study), Suaeda malacosperma (NC_039180), Chenopodium ficifolium (MK182725), Chenopodium quinoa (NC_034949, MK159176, MF805727, KY635884, and NSDK01003185), Chenopodium album (NC_034950 and MF418659), Spinacia oleracea (NC_002202), Dysphania pumilio (MH936550 and MK541016), Dysphania ambrosioides (MK182726), Beta vulgaris (EF534108), Beta vulgaris subsp. vulgaris (KJ081864), Beta vulgaris subsp. vulgaris ‘KWS2320’ (KR230391), Salicornia brachiate (NC_027224), Salicornia europaea (NC_027225), Salicornia bigelovii (NC_027226), Bienertia sinuspersici (KU726550), Haloxylon ammodendron (NC_027668), Haloxylon persicum (NC_027669), Amaranthus tricolor (KX094399), Amaranthus hypochondriacus (NC_030770 and MG836505), Amaranthus caudatus (NC_040143), Amaranthus hybridus subsp. cruentus (MG836506 and MG836507), Gymnocarpos przewalskii (NC_036812), Fagopyrum luojishanense (NC_037706). Phylogenetic tree was drawn based on neighbor joining tree. Right bars indicate each subfamily in the tree. The numbers above branches indicate bootstrap support values of neighbor joining and maximum likelihood phylogenetic trees, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Chung Y. 1992. A taxonomic study of the Korean Chenopodiaceae [PhD. Thesis]. Korea: Sungkyunkwan University (in Korean).
- Chung Y. 2018. Suaeda Forssk. ex J.F. Gmel. In: edited by Chong-wook Park Flora of Korea. Vol. 3. Caryophyllidae. Korea: Flora of Korean Editorial Committee, National Institute of Biological Resources.
- Devi RJ, Thongam B. 2017. Complete chloroplast genome sequence of Chenopodium album from Northeastern India. Genome Announc. 5:e01150–e01117.
- Hara H, Kurosawa S. 1963. Cytotaxonomical notes on some Japanese plants. J Jap Bot. 38:113–116.
- Hong S-Y, Cheon K-S, Yoo K-O, Lee H-O, Cho K-S, Suh J-T, Kim S-J, Nam J-H, Sohn H-B, Kim Y-H. 2017. Complete chloroplast genome sequences and comparative analysis of Chenopodium quinoa and C. album. Front Plant Sci. 8:1696.
- Hong S-Y, Cheon K-S, Yoo K-O, Lee H-O, Mekapogu M, Cho K-S. 2019. Comparative analysis of the complete chloroplast genome sequences of three Amaranthus species. Plant Genet Res. 1–10.
- Kadereit G, Borsch T, Weising K, Freitag H. 2003. Phylogeny of Amaranthaceae and Chenopodiaceae and the evolution of C4 photosynthesis. Int J Plant Sci. 164:959–986.
- Kapralov MV, Akhani H, Voznesenskaya EV, Edwards G, Franceschi V, Roalson EH. 2006. Phylogenetic relationships in the Salicornioideae/Suaedoideae/Salsoloideae sl (Chenopodiaceae) clade and a clarification of the phylogenetic position of Bienertia and Alexandra using multiple DNA sequence datasets. Syst Bot. 31:571–585.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim B, Kim J, Park H, Park J. 2016. The complete chloroplast genome sequence of Bienertia sinuspersici. Mitochondr DNA B. 1:388–389.
- Kim Y, Chung Y, Park J. 2019a. The complete chloroplast genome of Chenopodium ficifolium Sm. (Amaranthaceae). Mitochondrl DNA B. 4:872–873.
- Kim Y, Chung Y, Park J. 2019b. The complete chloroplast genome sequence of Dysphania pumilio (R. Br.) Mosyakin & Clemants (Amaranthaceae). Mitochondr DNA B. 4:403–404.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Maughan PJ, Chaney L, Lightfoot DJ, Cox BJ, Tester M, Jellen EN, Jarvis DE. 2019. Mitochondrial and chloroplast genomes provide insights into the evolutionary origins of quinoa (Chenopodium quinoa Willd.). Sci Rep. 9:185.
- Park J-S, Choi I-S, Lee D-H, Choi B-H. 2018. The complete plastid genome of Suaeda malacosperma (Amaranthaceae/Chenopodiaceae), a vulnerable halophyte in coastal regions of Korea and Japan. Mitochondr DNA B. 3:382–383.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
