Abstract
Nepeta cataria L. is the perennial herb and is a member of the Labiatae family, also has a considerable folkloric reputation in China. In this study, we presented, assembled, and annotated the complete chloroplast genome of N. cataria L. The chloroplast genome size is 153,526 bp, containing a large single-copy region (84,210 bp), a small single-copy region (18,464 bp), and a pair of IR regions (25,624 bp). The overall nucleotide composition is: 30.8% A, 31.1% T, 19.9% C, and 18.2% G, with a total AT content of the chloroplast genome 61.9% and GC content of 38.1%. Nepeta cataria L. whole chloroplast genome contains 134 genes, including 89 protein-coding genes (PCG), 37 transfer RNA (tRNAs), and 8 ribosome RNA (rRNAs). Phylogenetic Neighbor-Joining (NJ) analysis based on 24 herbal medicine plant species chloroplast genomes indicates that N. cataria L. is closely related to N. racemosa. This complete chloroplast genomes can be used for medicinal valuable species research of the Nepeta family.
Nepeta cataria L. is the most famous Nepeta species and the important herb plant in China. It is also a herbal medicinal treatment for fevers, diarrhea, insomnia, and lacking menstruation for humans (Herron Citation2003). It origins in China and later was introduced to Europe, America, and Southern Africa (Celenk et al. Citation2008). Rosmarinic acid is one of the main active constituents of Nepeta cataria L. and another important constituent is its aromatic oil. Nepeta cataria L. is a common food plant, which is a kind of high economic efficiency and has a great future and is a healthcare type vegetable. (Simona et al. Citation2015). However, our understanding of the chloroplast genome of the Nepeta cataria L. and the Nepeta family is limited. In order to further study the Nepeta cataria L. and the Nepeta family, genetic diversity and genetic structure of natural populations, we presented the complete chloroplast genome of Nepeta cataria L. and explore the phylogenetic relationship with other herbal medicine plants, which contributes to phylogenetic studies for these species and further conserve it.
The specimen sample of Nepeta cataria L. was isolated and deposited from Jiangxi Province Hospital of Integrated Chinese and Western Medicine (Nanchang, Jiangxi, China, 115.91E; 28.67N). Total genomic DNA was extracted from the blade tissue using Plant Tissues Genomic DNA Extraction Kit (Solarbio, BJ, CN) and stored in Jiangxi Province Hospital of Integrated Chinese and Western Medicine (No. JXPHICWM01). The total genomic DNA was used for the shotgun library construction and sequenced using the Illumina HiSeq X Ten Sequencing Platform (Illumina Co., San Diego, CA). Quality control was performed to remove low-quality reads and adapters using the FastQC software (Andrews Citation2015). The chloroplast genome was assembled using the Plann software (Huang and Cronk Citation2015) and annotated using the Geneious software (Simon et al. Citation2012). The physical map of the new chloroplast genome was generated using OGDRAW (Lohse et al. Citation2013). The accurate new annotated complete chloroplast genome was submitted to GenBank with accession number MH5654431.
The whole complete chloroplast genome of Nepeta cataria L. was a circle with 153,526 bp in size, containing a large single-copy region (LSC) of 84,210 bp, a small single- copy region (SSC) of 18,464 bp, and a pair of inverted repeat regions (IRA and IRB) of 25,624 bp. The cpDNA of N. cataria L. comprised 134 genes, including 89 protein-coding genes (PCG), 37 transfer RNA genes (tRNA), and 8 ribosomal RNA genes (rRNA). In the IR regions, a total of 19 genes were found duplicated, including 8 PCG species (rpl2, rpl23, ycf2, ycf15, ndhB, rps7, rps12, and ycf1), 7 tRNA species (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU) and 4 rRNA species (rrn16, rrn23, rrn4.5, and rrn5). The overall nucleotide composition is: 30.8% A, 31.1% T, 19.9% C, and 18.2% G, with a total AT content of 61.9% and GC content of 38.1%.
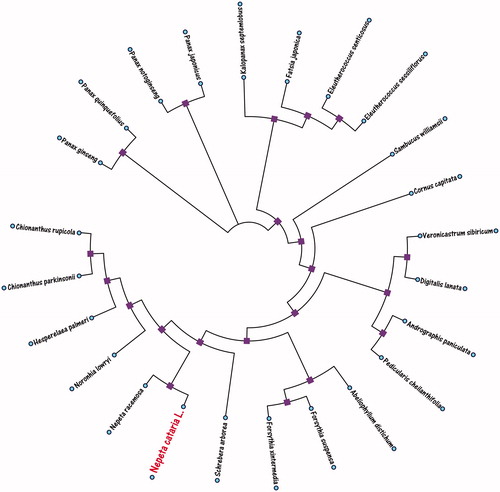
In Phylogenetic Neighbor-Joining (NJ) analysis, we selected other 23 herbal medicine plant species chloroplast genomes from GenBank to assess the relationship of Nepeta cataria L. The phylogenetic tree was reconstructed using neighbor-joining (NJ) methods. NJ analysis was performed using the MEGA X software with 5000 bootstrap values replicate (Kumar et al. Citation2018). The alignment was analyzed by the PhyloBayes v3.2 software (Lartillot et al. Citation2009) under the CATGTR + Γ model. All of the nodes were inferred with strong support by the NJ methods. The final tree was represented using the MEGA X software (Kumar et al. Citation2018) and edited using the iTOL Web software (Letunic and Bork Citation2016). As shown in the phylogenetic NJ tree result (), the chloroplast genome of Nepeta cataria L. is clustered and closest with Nepeta racemosa (No. MH626629.1) in the evolutionary relationship. However, the complete chloroplast of E. japonica is very important for the conservation and evolutionary studies of the Nepeta family.
Figure 1. The neighbor-joining (NJ) phylogenetic tree of the 24 species from Nepeta cataria L. was constructed based on the complete herbal medicine plants chloroplast genomes data. The analyzed herbal medicine species and corresponding GenBank accession numbers are as follows: Abeliophyllum distichum (KT274029.1), Andrographis paniculata (KF150644.2), Chionanthus parkinsonii (MG255752.1), Chionanthus rupicola (MG255753.1), Cornus capitata (MG524998.1), Digitalis lanata (KY085895.1), Eleutherococcus senticosus (JN637765.1), Eleutherococcus sessiliflorus (KT153019.1), Fatsia japonica (KR021045.1), Forsythia suspensa (MF579702.1), Forsythia xintermedia (MG255756.1), Hesperelaea palmeri (LN515489.1), Kalopanax septemlobus (KC456167.1), Nepeta racemosa (MH626629.1), Noronhia lowryi (MG255759.1), Panax ginseng (KF431956.1), Panax japonicus (KP036469.1), Panax notoginseng (KP036468.1), Panax quinquefolius (KT028714.1), Pedicularis cheilanthifolia (KY751712.1), Sambucus williamsii (KX510276.1), Schrebera arborea (MG255767.1), Veronicastrum sibiricum (KT724053.1).
Acknowledgments
No project pillar.
Disclosure statement
The authors have declared that no competing interests exist.
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Celenk S, Dirmenci T, Malyer H, Bicakci A. 2008. A palynological study of the genusnepetal. (lamiaceae). Plant Syst Evol. 276:105–123.
- Herron S. 2003. Catnip, Nepeta cataria, a morphological comparison of mutant and wild type specimens to gain an ethnobotanical perspective. Econ Bot. 57:135–142.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Lartillot N, Blanquart S, Lepage T. 2009. PhyloBayes 3.2: a Bayesian software for phylogenetic reconstruction and molecular dating using mixture models. Bioinformatics. 25:2286–2288.
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44:W242–W245.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Simon Buxton, Alex Cooper, Sidney Markowitz, Chris Duran, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- SimonaD, Liviu AM, Dan D, Otilia B. 2015. Overview regarding the bioactivity of agastache foeniculum and nepeta cataria species. Bull Univ Agric Sci Vet Med Cluj Napoca. 72:24–31.
