Abstract
Camellia renshanxiangiae is a vulnerable species distributed in China with unique hairy anthers and faint scent. Here, we report the characterization of its complete chloroplast (cp) genome using Illumina pair-end sequencing data. The total chloroplast genome size was 156,665 bp, including two inverted repeats (IRs) of 26,071 bp each, separated by a large single copy (LSC) and a small single copy (SSC) of 86,277 bp and 18,246 bp, respectively. A total of 131 genes, including 36 tRNA, 8 rRNA, and 87 protein-coding genes, were identified. Phylogenetic analysis showed that C. renshanxiangiae clustered with Camelia species (C. sinensis, C. crapnelliana).
The genus Camellia of the family Theaceae, comprising of a total of 200 species, has high economic and ornamental value. Approximately, 180 species of Camellia are distributed in south and southwest of China (Chang Citation1981). Camellia renshanxiangiae was first reported as a new species in 2001 (Ye and Zheng Citation2001). The presence of hairy anthers makes C. renshanxiangiae distinguishable from other species of the genus Camellia. In addition, small and white fragrant flowers are widely distributed in the branches (Song et al. Citation2009), a character that has been found in some rare species of this genus like C. lutchnensis, C. forrestii, C. grijsii, C. yunnanensis so far (Feng Citation1985). Currently, C. renshanxiangiae is listed as a vulnerable species globally (http://www.iucnredlist.org/details/62057348/0), thereby calling for efforts to conserve this valuable wild tea resource. A better insight into its genomics may contribute to its conservation. Towards this objective, we assembled and characterized the complete chloroplast genome of C. renshanxiangiae in this study. The annotated genomic sequence has been submitted to GenBank with the accession number MH253889.
Total DNA was isolated from fresh leaves of an individual of C. renshanxiangiae collected from Sun Yat-sen University, Guangzhou, Guangdong Province. The voucher specimen (Shi XG 171225) was deposited at the Sun Yat-sen University Herbarium (SYS). Genome sequencing was performed on an Illumina Hiseq X Ten (San Diego, California, U.S.) platform with paired-end reads of 150 bp. In total, 5.99 Gb short sequence data were obtained and used to assemble the chloroplast genome in NOVOPlasty (Dierckxsens et al. Citation2017). A partial chloroplast atpB sequence of a congeneric species Camellia leptophylla (GenBank accession number NC_024660) was used as the seed sequence. The genes in the chloroplast genome were annotated using the DOGMA program (Wyman et al. Citation2004). The circular chloroplast genome map was drawn using OGDRAW (Lohse et al. Citation2013).
The complete cp genome of C. renshanxiangiae was 156,665 bp in size, containing a pair of inverted repeats (IRs) of 26,071 bp each, which separated a large single copy region (LSC) of 86,277 bp and a small single copy region (SSC) of 18,246 bp (). The cp genome contained 131 genes, including 87 protein-coding genes, 36 transfer RNA genes, and 8 ribosomal RNA genes. Most of the genes occur as single-copy in the LSC or SSC, while 17 gene species had two copies in the IRs, including six protein-coding species, seven tRNA species, and all four rRNA species. The overall GC content of the cp genome was 37.33%.
Figure 1. Maximum-likelihood tree based on the sequences of 18 complete chloroplast genomes. Numbers in the nodes were bootstrap values from 1000 replicates. The position of Camellia renshanxiangiae is shown in bold and GenBank accession numbers are listed behind each species name.
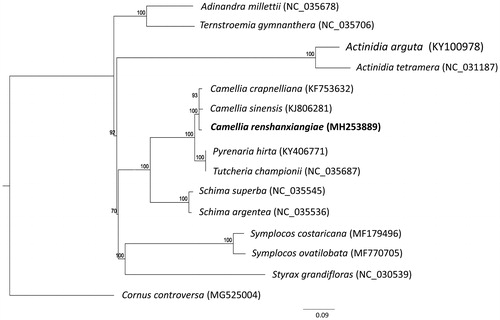
Phylogenomic analysis was performed using complete chloroplast genomes from 18 species in the order Ericales including C. renshanxiangiae and one species from Cornales (Cornus controversa) as the outgroup species. The chloroplast genome sequences were aligned using MAFFT (Katoh and Standley Citation2013). Phylogenetic analysis using the maximum likelihood algorithm was conducted with RAxML (Stamatakis Citation2014) implemented in Geneious ver. 10.1 (http://www.geneious.com, Kearse et al. Citation2012). The result showed that C. renshanxiangiae clustered with Camelia species (C. sinensis, C. crapnelliana) with 100% bootstrap support.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Chang HT. 1981. A taxonomy of the genus Camellia. Acta Sci Nat Univ Sunyatseni. 1:1–180.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Feng GM. 1985. Camellia flowers. World Agric. 2:48–51.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Steven SH, Matthew C, Shane S, Simon B, Alex C, Sidney M, Chris D, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucl Acids Res. 41:W575–W581.
- Song XH, Peng L, Shi XG, Dai ST, Ye CX. 2009. The aroma components from the flowers of Camellia renshanxiangiae by HS-SPME. Guihaia. 29:561–563.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Ye CX, Zheng XQ. 2001. A new species of the genus Camellia from Guangdong, China. Acta Phytotaxon Sin. 39:160–162.
