Abstract
In this study, the complete mitochondrial genome (mitogenome) sequence of Atrobucca nibe has been determined by polymerase chain reaction and primer walking methods. The mitogenome is a circular molecule of 16,504 bp in length, including the typical structure of 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and 2 non-coding regions (control region and L-strand replication origin), the gene contents of which are identical to those observed in most bony fishes. Within the control region, we identified the termination-associated sequence domain (TAS) and the conserved sequence block domain (CSB-F, CSB-E, CSB-D, CSB-C, CSB-B, CSB-A, CSB-1, CSB-2, CSB-3).
Atrobucca nibe belongs to the order Perciformes, the suborder Percoidei, the family Sciaenidae, and lives in the coastal waters down to a depth of 200 m or deeper. The species is an important economical fish and distributed in the Indo-West Pacific (Sasaki and Kailola Citation1988). Until now, there are some researches focused on its distribution and catching, but there are only a few studies on its genetic characteristics. Therefore, we determined the complete mitogenome sequence of A. nibe using 40 pairs of primers in order to provide basic information for the genetic studies of A. nibe.
The sample of A. nibe was collected from the coastal waters of Zhoushan of China during April 2017 and deposited in Fishery Ecology Laboratory of Ocean University of China. The complete mitogenome of A. nibe is sequenced to 16,504 bp and has high similarity on gene content and structure with most vertebrates (Cann et al. Citation1984; Boore Citation1999; Wolstenholme Citation1992). The mitogenome sequence has been deposited in GeneBank with accession number MK530652. It consists of 13 typical vertebrate protein-coding genes, 22 tRNA genes, 2 rRNA genes (12S rRNA and 16S rRNA), and 2 major non-coding regions (control region and L-strand replication origin). Most mitochondrial genes of A. nibe were encoded on the H-strand, while only ND6 and eight tRNA (Gln, Ala, Asn, Cys, Tyr, Ser-UCN, Glu, and Pro) genes encoded on the L-strand. The A. nibe mitochondrial genes overlapped by a total of 29 bp, most of which occurred in protein-coding genes (ATPase6-ATPase8, ND4-ND4L). The overlap of the ATPase genes was 10 bp. Overall, base compositions of A. nibe mitochondrial genome were as follows: C, 31.01%; A, 27.95%; T, 24.77%; G, 16.27%, indicating a strand compositional bias characterized by a excess of C relative to G. The total length of 13 protein-coding genes was 11,435 bp, accounting for 69.29% of the complete mitogenome sequences, and contained 3802 amino acids for protein coding. Like other mitochondrial genomes, 22 tRNA genes of A. nibe mitochondrial genome were detected and interspersed between the rRNA and protein-coding genes and ranged in size from 66 to 74 bp (Cheng et al. Citation2012; Zhao et al. Citation2012; Yang et al. Citation2016). A small subunit of rRNA (12S rRNA) and a large subunit of rRNA (16S rRNA) were also identified in A. nibe mitogenome, which were 950 and 1695 bp, respectively. The largest non-coding region (control region) in A. nibe mitogenome, which was located between tRNAPro and tRNAPhe, was determined to be 825 bp in length. By comparing with the recognition sites in some other bony fishes, the control region was characterized by the typical tripartite with termination-associated sequence domain, the central conserved sequence block domains (CSB-F, CSB-E, CSB-D, CSB-C, and CSB-B, and CSB-A), and the CSB domains (CSB-1, CSB-2 and CSB-3) (Cui et al. Citation2009; Cheng et al. Citation2010; Liu et al. Citation2016). The additional non-coding region, the origin of L-strand replication (OL) in A. nibe, was 38 bp in size and was located between tRNAAsn and tRNACys in the WANCY region, almost identical with other Sciaenidae fishes. The putative OL can form a stable stem-loop secondary structure with the stem formed by 14 paired nucleotides and the loop of 13 nucleotides. We expect that the present result would facilitate the resolution of taxonomic confusion and be helpful to understand the evolutionary history of A. nibe.
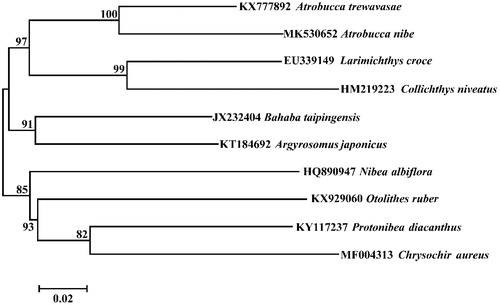
Only one complete mitochondrial genome sequence of other Atrobucca species, Atrobucca trewavasae has been found in NCBI. Phylogenetic relationships were constructed using the NJ algorithm among these two Atrobucca species and some Sciaenidae fishes based on 12 H-strand mitochondrial protein-coding genes, 22 tRNA and 2 rRNA genes (). This phylogenetic tree shows that A. nibe is more closely related to Atrobucca trewavasae and more far related to other Sciaenidae species.
Disclosure statement
The authors declare that there are no conflicts of interest and authors are solely responsible for the contents and writing of the paper.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Cann RL, Brown WM, Wilson AC. 1984. Polymorphic sites and the mechanism of evolution in human mitochondrial DNA. Genetics. 106:479–499.
- Cheng YZ, Xu TJ, Shi G, Wang RX. 2010. Complete mitochondrial genome of the miiuy croaker Miichthys miiuy (Perciformes, Sciaenidae) with phylogenetic consideration. Mar Genomics. 3:201–209.
- Cheng YZ, Xu TJ, Jin XX, Shi G, Wang RX. 2012. The complete mitochondrial genome of silver croaker Argyrosomus argentatus (Perciforems; Sciaenidae): genome characterization and phylogenetic consideration. Mol Biol. 46:224–233.
- Cui ZX, Liu Y, Li CP, You F, Chu KH. 2009. The complete mitochondrial genome of the large yellow croaker, Larimichthys crocea (Perciformes, Sciaenidae): unusual features of its control region and the phylogenetic position of the Sciaenidae. Gene. 432:33–43.
- Liu L, Yang H, Yang Z, Zhao H, Sun J, Xiao S, Yang X, Li G. 2016. The complete mitochondrial genome of the blackspotted croaker Protonibea diacanthus (Perciformes, Sciaenidae). Mitochondr DNA A. 27:1671–1673.
- Sasaki K, Kailola PJ. 1988. Three new Indo-Australian species of the sciaenid genus Atrobucca, with a reevaluation of generic limit. Jap J Ichthyol. 35:261–277.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216.
- Yang H, Xie Z, Li S, Wu X, Peng C, Zhang Y, Lin H. 2016. The complete mitochondrial genome of the orange-spotted grouper Epinephelus coioides (Perciformes, Serranidae). Mitochondr DNA A. 27:1674–1676.
- Zhao LL, Gao TX, Lu WH, Lu NN, Li YZ, Zhang ZH. 2012. The complete mitogenome of the Chinese bahaba Bahaba taipingensis (Perciformes: Sciaenidae). Mitochondr DNA. 23:411–413.