Abstract
Artemisia fukudo Makino is a salt tolerant species and contains diverse useful compounds. Here, we completed chloroplast genome of A. fukudo to understand intraspecies variations producing better useful compounds. Its length is 151,021 bp long and has four subregions: 82,761 bp of large single copy (LSC) and 18,348 bp of small single copy (SSC) regions are separated by 24,956 bp of inverted repeat (IR) regions including 125 genes (88 protein-coding genes, eight rRNAs, and 37 tRNAs). The overall GC content of this chloroplast genome is 37.5% and those in the LSC, SSC, and IR regions are 35.6, 30.7, and 43.1%, respectively. Seven single nucleotide polymorphisms and 11 insertions and deletions are identified between A. fukudo isolated in different islands in the same county. Phylogenetic trees show that two A. fukudo are clustered in one clade. In addition, phylogenetic position of Artemisia annua is different from the previous study caused by lack of enough taxa. This chloroplast genome will contribute understanding of both intraspecies variations of A. fukudo andmolecular phylogeny of Artemisia genus.
The genus Artemisia is one of the largest and widely distributed genera in family Asteraceae (Compositae) (Bremer and Anderberg Citation1994; Watson et al. Citation2002), covering more than 500 species mainly distributed in the temperate zones of Europe, Asia, and North America (Shreve Citation1942; Ling Citation1990; Ling Citation1991; Young et al. 1996). Artemisia fukudo Makino is distributed in the seashore of Korea and western Japan (Ishikawa and Kachi Citation2000). Due to the coastal habitats, A. fukudo is salt tolerant (Ishikawa and Kachi Citation2000). Artemisia fukudo also has useful features, such as its essential oil attenuates inflammation (Yoon et al. Citation2007; Yoon et al. Citation2010), anticancer activities (Kim et al. Citation2007; Emami et al. Citation2009), and anticomplementary activity (Moon et al. Citation2012). In different habitats and/or genetic background, densities of useful compounds among plants are different (Keskitalo et al. Citation2001; Montoro et al. Citation2013).
To investigate intraspecies variations which can produce more useful compounds, total DNA of A. fukudo isolated from Aphae-do, Shinan-gun, Jeollanam-do, Korea (Voucher in InfoBoss Cyber Herbarium (IN); Y. Kim, IB-00595) was extracted from fresh leaves by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome was sequenced using HiSeqX at Macrogen Inc., Seoul, Korea, and de novo assembly and confirmation of assembled sequence were performed by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for annotation based on previously published A. fukudo chloroplast genome (KU360270; Lee et al. Citation2016).
The chloroplast genome (Genbank accession is MK569048) is 151,021 bp (GC ratio is 37.5%) and has four subregions: 82,761 bp of large single copy (LSC; 35.6%) and 18,348 bp of small single copy (SSC; 30.7%) regions are separated by 24,956 bp of inverted repeat (IR; 43.1%). It contains 125 genes (88 protein-coding genes, eight rRNAs, and 37 tRNAs); 18 genes (seven protein-coding gene, four rRNAs, and seven tRNAs) are duplicated in IR regions.
Seven single nucleotide polymorphisms (SNPs) and eleven insertions and deletions are identified between Jeung-do and Aphae-do isolates, similar to samples in different countries (Park et al. Citation2019). Two non-synonymous SNPs are found in rps7, one synonymous SNP is identified in rps15, and one insertion is in psbT.
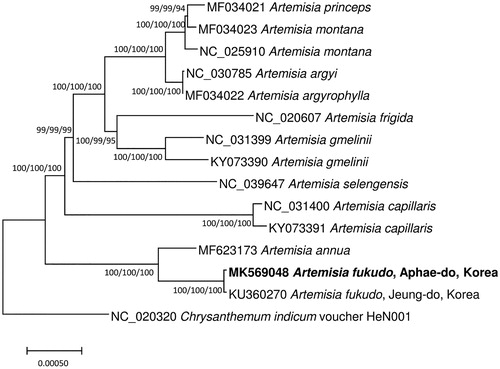
Fourteen Artemisia and one Chrysanthemum chloroplast genomes as outgroup species (Watson et al. Citation2002) were used for constructing bootstrapped neighbor joining, minimum evolution, and maximum likelihood phylogenic trees using MEGA X (Kumar et al. Citation2018) after aligning whole chloroplast genomes with inverting 82 kb inversion of Atemisia annua (Shen et al. Citation2017) and SSC regions in three genomes (A. annua, Artemisia princeps, and Chrysanthemum indicum) using MAFFT 7.388 (Katoh and Standley Citation2013). Phylogenetic trees show that two A. fukudo are clearly clustered as expected (). In addition, phylogenetic position of A. annua in phylogenetic trees is different from previous study (Watson et al. Citation2002) due to insufficient taxa to recover phylogenetic relation of Atemisia species (). This chloroplast genome will contribute understanding of intraspecies variation of A. fukudo and Artemisia phylogeny.
Figure 1. Neighbor joining (bootstrap repeat is 10,000), minimum evolution (bootstrap repeat is 10,000), and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of fourteen Artemisia and one Chrysanthemum: Artemisia fukudo (MK569048 in this study and KU360270), Artemisia gmelinii (NC_031399 and KY073390), Artemisia selengensis (NC_039647), Artemisia argyi (NC_030785), Artemisia argyrophylla (MF034022), Artemisia princeps (MF034021), Artemisia montana (MF034023 and NC_025910), Artemisia frigida (NC_020607), Artemisia capillaris (NC_031400 and KY073391), Artemisia annua (MF623173), and Chrysanthemum indicum (NC_020320). Phylogenetic tree was drawn based on neighbor joining tree. The numbers above branches indicate bootstrap support values of neighbor joining, minimum evolution, and maximum likelihood phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bremer K, Anderberg AA. 1994. Asteraceae: cladistics and classification. Sirsi. (i9780881922752).
- Emami SA, Vahdati-Mashhadian N, Vosough R, Oghazian MB. 2009. The anticancer activity of five species of Artemisia on Hep2 and HepG2 cell lines. Pharmacologyonline. 3:327–339.
- Ishikawa SI, Kachi N. 2000. Differential salt tolerance of two Artemisia species growing in contrasting coastal habitats. Ecol Res. 15:241–247.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Keskitalo M, Pehu E, Simon JE. 2001. Variation in volatile compounds from tansy (Tanacetum vulgare L.) related to genetic and morphological differences of genotypes. Biochem Syst Ecol. 29:267–285.
- Kim K-N, Lee J-A, Yoon W-J, Kim J-Y, Song G-P, Park S-Y. 2007. The cytotoxicity of Artemisia fukudo extracts against HL-60 cells. J Korean Soc Food Sci Nutr. 36:819–824.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Lee YS, Park JY, Kim J-K, Lee HO, Park H-S, Lee S-C, Kang JH, Lee TJ, Sung SH, Yang T-J. 2016. Complete chloroplast genome sequence of Artemisia fukudo Makino (Asteraceae). Mitochondr DNA B. 1:376–377.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Ling Y. 1990. Hengduang-Himalayan Mountains (HH), a special area from the floristic point of view for Artemisia L.(Compositae). Bull Bot Res. 10:73–92.
- Ling Y-R. 1991. The Old World Artemisia (Compositae). Bull Bot Lab N-East for Inst. 12:1–108.
- Management of woody species of Artemisia in western North America. 1996. Compositae: Biology and Utilization Proceedings of the International Compositae Conference 1994. In Caligari P, Hind D, editors. Kew: Royal Botanic Gardens.
- Montoro P, Molfetta I, Maldini M, Ceccarini L, Piacente S, Pizza C, Macchia M. 2013. Determination of six steviol glycosides of Stevia rebaudiana (Bertoni) from different geographical origin by LC-ESI-MS/MS. Food Chem. 141:745–753.
- Moon H-I, Jung S, Lee Y-C, Lee J-H. 2012. Anticomplement activity of various solvent extracts from Korea local Artemisia spp. Immunopharmacol Immunotoxicol. 34:95–97.
- Park J, Kim Y, Kwon W, Nam S, Song MJ. 2019. The second complete chloroplast genome sequence of Nymphaea alba L. (Nymphaeaceae) to investigate inner-species variations. Mitochondr DNA B. 4:1014–1015.
- Shen X, Wu M, Liao B, Liu Z, Bai R, Xiao S, Li X, Zhang B, Xu J, Chen S. 2017. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules. 22:1330.
- Shreve F. 1942. The desert vegetation of North America. Bot Rev. 8:195–246.
- Watson LE, Bates PL, Evans TM, Unwin MM, Estes JR. 2002. Molecular phylogeny of subtribe Artemisiinae (Asteraceae), including Artemisia and its allied and segregate genera. BMC Evol Biol. 2:17.
- Young J, Longland W, Blank R. Management of woody species of Artemisia in western North America. Compositae: Biology and Utilization Proceedings of the International Compositae Conference 1994 (Edited by: P Caligari, D Hind) Royal Botanic Gardens, Kew; 1996.
- Yoon W-J, Lee J-A, Kim K-N, Kim J-Y, Park S-Y. 2007. In vitro anti-inflammatory activity of the Artemisia fukudo extracts in murine macrophage RAW 264.7 cells. Korean J Food Sci Technol. 39:464–469.
- Yoon W-J, Moon J, Song G, Lee Y, Han M, Lee J, Ihm B, Lee W, Lee N, Hyun C. 2010. Artemisia fukudo essential oil attenuates LPS-induced inflammation by suppressing NF-kappaB and MAPK activation in RAW 264.7 macrophages . Food Chem Toxicol. 48:1222–1229.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Gen Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
