Abstract
The nearly complete mitogenome of aquatic predator species, Lispe assimilis Wiedemann, 1824, was obtained using a next-generation sequencing approach, which is the first report in Coenosiinae (Calyptratae, Muscidae). The genome is 15,386 bp long with high A + T content. The phylogenetic analyses based on 13 protein-coding genes (PCGs) and 2 ribosomal RNA (rRNA) genes provide strong support for monophyly of the Muscidae, confirming divergence of Coenosiinae from the related subfamilies (Azeliinae and Mydaeinae).
The genus Lispe Latreille, 1979 belongs to Coenosiinae (Diptera: Calyptratae, Muscidae), distributes in all zoogeographical regions with the exception of New Zealand (Vikhrev Citation2015). As active predators inhabited in semi-aquatic environments, the larvae or adults of Lispe feed on a variety of pest groups such as mosquitos, making them the indicators of water quality (Xue & Zhang Citation2005; Ge et al. Citation2016). Here, we document the nearly complete mitogenome of this group for the first time and present the phylogeny of the Muscidae, which provides a hypothesis on the evolution of Muscidae from mitogenomic perspective.
A male pinned specimen of Lispe assimilis Wiedemann, 1824 was collected on 8 July 2013 from Hubei Province, China (N29°43′48″, E112°24′36″) and deposited in the Museum of Beijing Forestry University, Beijing with an accession number L01410. The genomic DNA was extracted with phenol-chloroform as described in Ge et al. (Citation2016) without crashing specimen. The genomic DNA was pooled with other insect species and sequenced using the Illumina HiSeq2500 (PE250, Illumina, San Diego, CA) platform (Gillett et al. Citation2014). In total, 2.66 G raw reads were obtained and trimmed using Trimmomatic (Bolger et al. Citation2014). Subsequently, DNA reads were assembled employing idba_ud implemented with IDBA-1.1.1 (Peng et al. Citation2012). Mitogenome was pulled out with COI as bait sequence (Ding et al. Citation2019) and annotated referring Zhang et al. (Citation2016). Phylogenetic tree based on 13 protein-coding genes (PCGs) and two ribosomal RNA (rRNA) genes were inferred using Maximum Likelihood (ML) and Bayesian algorithms as described in Yan et al. (Citation2017).
The nearly complete mitogenome is 15,385 bp in length (GenBank accession No. MK607086), containing one non-coding control region and 37 canonical mitochondrial genes. Among the 13 PCGs, only ATP8 uses start codon ATC, the rest are encoded by the typical ATT (ND2, COX1, ND5, ND6, CYTB), ATG (COX2, ATP6, COX3, ND4), and ATA (ND3, ND4L, ND1) start codons. Particularly, an incomplete stop codon T was found in COX1 and COX2, while the rest were encoded by TAA stop codons. The overall nucleotide composition was estimated to be 40.08% of A, 39.60% of T, 12.22% of C, and 8.10% of G, with a slightly higher A + T content (79.68%) than other calyptratae (Zhang et al. Citation2016).
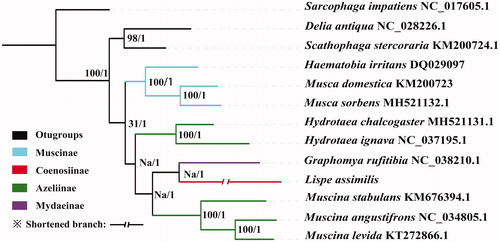
The monophyly of Muscidae is well-supported in both ML and Bayesian analyses (posterior probabilities = 1, bootstrp= 100) which is in agreement with previous studies () (Kutty et al. Citation2014 ). The Muscinae is also confirmed as monophyletic based on our phylogenetic hypotheses. Otherwise, Coenosiinae is nested in Azeliinae with low support (bootstrp = 31) in ML analyses, while the Bayesian tree shows that the Coenosiinae species Lispe assimilis is sister to Mydaeinae (posterior probabilities = 1).
Acknowledgments
The authors are grateful to Prof. Hu Li (China Agricultural University), Mr. Fan Song (China Agricultural University) and Mr. Zhenyong Du (China Agricultural University) for their help in bioinformatics analysis. We thank Mr. Ming Zhang (Beijing Forestry University) for specimen collection and Mr. Yingqiang Ge (Beijing Forestry University) for specimen identification.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Ding SM, Zhao CJ, Yang D. 2019. Complete mitochondrial genome of Heterostomus Sp. (Diptera, Xylophagidae). Mitochondrial DNA B. 4:884–885.
- Ge YQ, Gao YY, Yan LP, Liu XH, Zhang D. 2016. Review of the Lispe tentaculata-group (Diptera: Muscidae) in China, with one new synonym. Zoosystema. 38:339–352.
- Gillett C, Crampton-platt A, Timmermans M, Jordal BH, Emerson BC, Vogler AP. 2014. Bulk de novo mitogenome assembly from pooled total DNA elucidates the phylogeny of weevils (Coleoptera: Curculionoidea). Mol Biol Evol. 31:2223–2237.
- Kutty SN, Pont AC, Meier R, Pape T. 2014. Complete tribal sampling reveals basal split in Muscidae (Diptera), confirms saprophagy as ancestral feeding mode, and reveals an evolutionary correlation between instar numbers and carnivory. Mol Phylogenet Evol. 78:349–364.
- Peng Y, Leung HC, Yiu SM, Chin FY. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28:1420–1428.
- Vikhrev N. 2015. Taxonomy notes on Lispe (Diptera, Muscidae). Parts 10–12. Amu Zool J. 7:228–247.
- Xue WQ, Zhang D. 2005. A review of the genus Lispe Latreille (Diptera: Muscidae) from China, with descriptions of new species. Orient Insects. 39:117–139.
- Yan LP, Zhang M, Gao YY, Thomas P, Zhang D. 2017. First mitogenome for the subfamily Miltogramminae. (Diptera: Sarcophagidae) and its phylogenetic implications. Eup J Entomol. 114:442–449.
- Zhang D, Yan LP, Zhang M, Chu HJ, Cao J, Li K, Hu DF, Thomas P. 2016. Phylogenetic inference of calyptrates, with the first mitogenomes for Gasterophilinae (Diptera: Oestridae) and Paramacronychiinae (Diptera: Sarcophagidae). Int J Biol Sci. 12:489–504.