Abstract
The complete mitochondrial genome of the three-keeled pond turtle Mauremys reevesii from Japan was analyzed using next-generation sequencing. The mitochondrial genome of M. reevesii was observed to be a circular molecule of 15,528 bp containing 13 protein-coding genes, 22 tRNA genes, and two rRNA genes, along with one A + T-rich control region. The average AT was found to be 61.11%. One extra base was present at the 174th position in the ND3 gene. Molecular phylogenetic analysis revealed that M. reevesii from Japan was few genetically distant from the geographically isolated M. reevesii from South Korea. This turtle might have been exported to Japan from the Korean Peninsula after the Middle Ages.
The three-keeled pond turtle Mauremys reevesii is distributed across China, Taiwan, the Korean Peninsula, and Japan (Lovich et al. Citation1985). Mauremys reevesii inhabits main islands of Hokkaido, Honshu, Shikoku, Kyushu, and Okinawa, as well as some remote islands of Japan (Suzuki and Hikida Citation2011). However, M. reevesii in Japan and Taiwan is thought to have been introduced from these areas after the Middle Ages according to some ancient documents (Suzuki et al. Citation2014). Here, we report the complete mitochondrial genome of the three-keeled pond turtle M. reevesii from Japan. The obtained data can be used to determine whether this turtle is a native species or a foreign species.
DNA samples from oral cells of M. reevesii collected from ponds in Kyoto City, Japan (35°04′08.8″N, 135°45′23.8″E) were immediately extracted using DNeasy mini kit (QIAGEN) for mitochondrial DNA analysis. The specimen was stored in a freezer at −20 °C in our laboratory at the Kyoto Sangyo University (Japan).
Genomic DNA isolated from oral cells was sequenced using Illumina’s MiSeq platform (Illumina). The complete mitochondrial genome of the Korean M. reevesii was used as a reference sequence (KJ700438). The resultant reads were assembled and annotated using the MITOS web server (Bernt et al. Citation2013) and Geneious R9 (Biomatters) (Kearse et al. Citation2012).
We succeeded in sequencing the entire mitochondrial genome of M. reevesii from Kyoto (available from DDBJ accession numbers AP019398). The genomes consisted of a 15,528 bp closed loop containing 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and one AT-rich control region, which represents a typical turtle mitochondrial genome (Shin et al. Citation2015; Zhao, Zhang, et al. Citation2016a; Zhao, Li, et al. Citation2016b; Li et al. Citation2017; Feng et al. Citation2017; Wang et al. Citation2017). The average AT content of the M. reevesii mitochondrial genome was 61.11%. The heavy strand was predicted to have 12 PCGs and 14 tRNA genes, whereas the light strand was predicted to contain one PCG and eight tRNA and two rRNA genes. PCGs began with ATG as the start codon, except for COI, with GTG as the start codon. Twelve PCGs used TAA as the stop codon, whereas only COI used AGG as the stop codon. Incomplete stop codons TA (found in COIII and Cytb) were identified. One extra base was present at the 174th position in the ND3 gene. This was similar to the results of turtles in previous studies (Mindell et al. Citation1998).
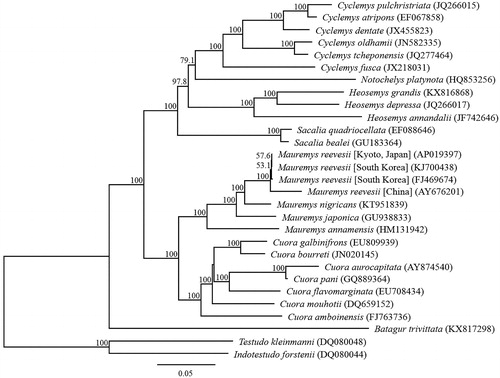
Phylogenetic analysis was performed using TREEDINDER (Jobb Citation2011) based on nucleotide sequences of 13 PCGs and the corresponding PCG of 29 closely related taxa (). The complete mitochondrial genomes of M. reevesii from Japan were few different from those of South Korea. This result is also consistent with the value of genetic distance, which is generally observed among the partial mitochondrial genomes of M. reevesii (Honda et al. Citation2002; Fong and Chen Citation2010; Suzuki et al. Citation2011; Guillon et al. Citation2012; Oh et al. Citation2017). This turtle might have been exported to Japan from the Korean Peninsula after the Middle Ages.
Figure 1. Phylogenetic relationships (maximum likelihood) of pond turtles based on nucleotide sequences of the 13 protein-coding genes of the mitochondrial genome. The numbers at the nodes indicate bootstrap support inferred from 1,000 bootstrap replicates. Alphanumeric terms in parentheses indicate the DNA Database of Japan accession numbers.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Feng L, Yang J, Zhang YP, Zhao GF. 2017. The complete mitochondrial genome of the Burmese roofed turtle (Batagur trivittata) (Testudines: Geoemydidae). Conserv Genet Resour. 9:95–97.
- Fong JJ, Chen TH. 2010. DNA evidence for the hybridization of wild turtles in Taiwan: possible genetic pollution from trade animals. Conserv Genet. 11:2061–2066.
- Guillon JM, Guéry L, Hulin V, Girondot M. 2012. A large phylogeny of turtles (Testudines) using molecular data. Contributions Zool. 81:147–158.
- Honda M, Yasukawa Y, Hirayama R, Ota H. 2002. Phylogenetic relationships of the Asian box turtles of the genus Cuora sensu lato (Reptilia: Bataguridae) inferred from Mitochondrial DNA sequences. Zool Sci. 19:1305–1312.
- Jobb G. 2011. TREEFINDER version of March 2011. Munich. http://www.treefnder.de.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Li J, Lu Y, Zan J, Nie L. 2017. Complete mitochondrial genome of the Cyclemys pulchristriata (Chelonia: Geoemydidae). Mitochondrial DNA Part B. 2:403–404.
- Lovich JE, Ernst CH, Gotte SW. 1985. Geographica variation in the Asiatic turtle Chinemys reevesii (Gray) and the status of Geoclemys granger Schmidt. J Herpetol. 19:238–245.
- Mindell DP, Michael D, Sorenson MD, Dimcheff DE. 1998. An extra nucleotide is not translated in mitochondrial ND3 of some birds and turtles. Mol Biol Evol. 15:1568–1571.
- Oh HS, Park SM, Han SH. 2017. Mitochondrial haplotype distribution and phylogenetic relationship of an endangered species Reeve’s turtle (Mauremys reevesii) in East Asia. J Asia Pac Biodiv. 10:27–31.
- Shin HW, Jang KH, Ryu SH, Choi HE, Hwang UW. 2015. Complete mitochondrial genome of the Korean Reeves’s turtle Mauremys reevesii (Reptilia, Testudines, Geoemydidae). Mitochondrial DNA Part A. 26:676–677.
- Suzuki D, Hikida T. 2011. Mitochondrial phylogeography of the Japanese pond turtle, Mauremys japonica (Testudines, Geoemydidae). J Zool Syst Evol Res. 49:141–147.
- Suzuki D, Ota H, Oh HS, Hikida T. 2011. Origin of Japanese populations of Reeves' pond turtle, Mauremys reevesii (Reptilia: Geoemydidae), as inferred by a molecular approach. Chelonian Conserv Biol. 10:237–249.
- Suzuki D, Yabe T, Hikida T. 2014. Hybridization between Mauremys japonica and Mauremys reevesii inferred by Nuclear and Mitochondrial DNA Analyses. J Herpetol. 48:445–454.
- Wang Y, Dai X, Wang M, Nie L. 2017. The complete mitochondrial genome of the Heosemys depressa (Testudines, Geoemydidae). Mitochondrial DNA Part B. 2:437–438.
- Zhao J, Zhang X, Li W, Zhang D, Zhu X. 2016a. The complete mitochondrial genome of the endangered Chinese black-necked pond turtle, Mauremys nigricans. Mitochondrial DNA Part B. 1:64–65.
- Zhao J, Li W, Zhang D, Wen P, Zhu X. 2016b. The mitochondrial genomes of three lineages of Asian yellow pond turtle, Mauremys mutica. Mitochondrial DNA Part A. 27:2466–2467.
