Abstract
This study reports the complete mitochondrial sequence of the Roman snail, one of the largest terrestrial snails of Europe. Two specimens of Helix pomatia were sequenced, which had a sequence length of 14,070 and 14,072 base pairs. The mitogenome has the common metazoan makeup (13 protein-coding genes, two ribosomal RNAs, and 21 transfer RNAs) and the ‘typical’ Helicid gene order (Cytochrome Oxidase subunit III and tRNA-T translocated compared to other Helicoidea). All protein coding genes could be annotated with proper start and stop codons (no completion of stop codons by polyadenylation). The specimens have a sequence similarity of 99.0% (146 differences) and the average base composition is 29.7% A, 15.0% C, 17.9% G, and 37.4% T. Phylogenetic anlyses with available Helicid mitogenomes show Helicinae as monophyly and H. pomatia as the sister-group of (Cepaea nemoralis, Theba pisana, and Cornu aspersum).
Helix pomatia is indigenous to Europe but has also been introduced in Asia and America. It is the type species of the genus. The species was recently split in H. pomatia and H. thesallica (Korábek et al. Citation2016). The former is shown to consist of multiple lineages, of which the specimens analyzed in this study correspond to the widespread clade A in (Korábek et al. Citation2018). Both specimens were collected (15.V.2015) in Aisne (Hirson, Blangy, France) at 49.942762 N 4.087542 E and stored in absolute ethanol. Vouchers were deposited in the collection of Naturalis Biodiversity Center, Leiden (RMNH.5006707.1 and .2). DNA was extracted with a DNeasy Tissue Kit (Qiagen). Partial Cytochrome Oxidase subunit I (COXI) and Cytochrome B (CYTB) sequences were amplified and Sanger sequenced with previously published universal primers (Folmer et al., Citation1994; Merritt et al., Citation1998). Based on these sequences, Long Range (LR) PCR primers were designed to amplify the mitogenome, using GoTaq long PCR Master Mix (Promega), in two fragments (sizes 6399 and 7592 nt):
Hel_pom_A_F5′-TTTATCCACCTCTTAGCAGTTTAATTGG-3′
Hel_pom_A_R5′-AAAATATGGGTGGAAAACGATCTTAGAG-3′ and
Hel_pom_B_F5′-TCCTCTTGGTAATCTTAATCATGTCTCT'-3'
Hel_pom_B_R5′-CTTATATCAGGTGCACCAATAAGTAAGG-3′
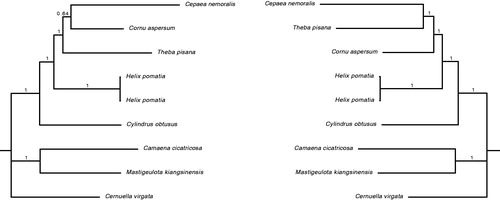
The H. pomatia LR PCR products were part of a combined run. Products were pooled equimolar and labeled per specimen. Library prep was done as described in (Groenenberg et al. Citation2017). The library was paired end sequenced on an Illumina Miseq at Macrogen Inc. (Korea). The run resulted in 790,034 and 982,448 reads for RMNH.5006707.1 and .2, respectively. The mitogenomes were constructed using a de Novo assembly and annotated by mapping against (in particular) Cornu (Gaitán-Espitia et al. Citation2013) and Cylindrus (Groenenberg et al. Citation2012). Average nucleotide position coverages were 10842 (sd 5288) and 15920 (sd 5053) times. The location of the tRNAs was confirmed with ARWEN (Laslett and Canback Citation2008). The start codon ATG was used six times (COX1, ND5, COX2, ATP8, ATP6, COX3), GTG three times (ND4L, ND4, ND2), TTG two times (ND6, ND3), ATA one time (ND1), and ATC one time (CYTB). The stop codon TAA was used eight times (COX1, ND6, ND5, CYTB, ATP6, ND3, COX3, ND4) and TAG five times (ND1, ND4L, COX2, ATP8, ND2). Controversy exists regarding the relationships within Helicinae. Either H. pomatia together with Cepaea nemoralis is the sister-group of Cornu aspersum and Theba pisana (Razkin et al. Citation2015), or H. pomatia is the sister-group of the latter three (Neiber and Hausdorf Citation2015). Separate MrBayes v.3.2.6 (Ronquist and Huelsenbeck Citation2003) analyses for both nucleotide (based on 41 partitions as in) and amino acid (model selection with PartitionFinder v.2.1.1. (Lanfear et al., Citation2016)) datasets confirm the monophyly of (C. nemoralis, C. aspersum, and T. pisana) to the exclusion of H. pomatia (). Relationships within the latter clade cannot be established with the current dataset.
Figure 1. Bayesian phylogenies based on nucleotide (left) and amino acid (right) alignments of complete mitochondria currenlty available for Helicidae (GenBank accession numbers: NC_001816, NC_017872, NC_021747 and MH362760). The tree is rooted with representatives of Geomitridae (NC_030723), Bradybaenidae (NC_024935) and Camaenidae (NC_025511).
The sequences of this study have been deposited in GenBank under accession numbers MK488030 and MK488031.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Marine Biol Biotechnol. 3:294–299.
- Gaitán-Espitia JD, Nespolo RF, Opazo JC. 2013. The complete mitochondrial genome of the land snail Cornu aspersum (Helicidae: Mollusca): intra-specific divergence of protein-coding genes and phylogenetic considerations within Euthyneura. PLoS One. 8:e67299.
- Groenenberg DSJ, Harl J, Duijm E, Gittenberger E. 2017. The complete mitogenome of Orcula dolium (Draparnaud, 1801); ultra-deep sequencing from a single long-range PCR using the Ion-Torrent PGM. Hereditas. 154:7
- Groenenberg DSJ, Pirovano W, Gittenberger E, Schilthuizen M. 2012. The complete mitogenome of Cylindrus obtusus (Helicidae, Ariantinae) using Illumina next generation sequencing. BMC Genomics. 13:114.
- Korábek O, Juřičková L, Petrusek A. 2016. Splitting the Roman snail Helix pomatia Linnaeus, 1758 (Stylommatophora: Helicidae) into two: Redescription of the forgotten Helix thessalica Boettger, 1886. J Mollusc Stud. 82:11–22.
- Korábek O, Petrusek A, Juřičková L. 2018. Glacial refugia and postglacial spread of an iconic large European land snail, Helix pomatia (Pulmonata: Helicidae). Biological Journal of the Linnean Society. 123:218–234.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34:msw260.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Merritt TJ, Shi L, Chase MC, Rex M. a, Etter RJ, Quattro JM. 1998. Universal cytochrome b primers facilitate intraspecific studies in molluscan taxa. Mol Marine Biol Biotechnol. 7:7–11.
- Neiber MT, Hausdorf B. 2015. Molecular phylogeny reveals the polyphyly of the snail genus Cepaea (Gastropoda: Helicidae). Mol Phylogenet Evol. 93:143–149.
- Razkin O, Gómez-Moliner BJ, Prieto CE, Martínez-Ortí A, Arrébola JR, Muñoz B, Chueca LJ, Madeira MJ. 2015. Molecular phylogeny of the western Palaearctic helicoidea (Gastropoda, Stylommatophora). Mol Phylogenet Evol. 83:99–117.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
