Abstract
Cardiocondyla obscurior is native to Southeast Asia but has become successfully established throughout the tropics and subtropics. Here, its mitochondrial genome was assembled from Illumina sequencing data. The genome is 15,213 bp long with a highly asymmetric nucleotide composition, and harbors the typical set of 37 mitochondrial genes and one noncoding control region. All PCGs are initiated with typical ATN codons and are terminated with TAA, TAG or the incomplete T(aa) codon. The control region has a remarkably high A + T content (90.8%) and harbors a 34-bp-long tandem repeat (TA)17. Phylogenetic analysis suggests that it is closely related to Pristomyrmex punctatus.
The tramp ant Cardiocondyla obscurior is native to Southeast Asia but has become successfully established throughout the tropics and subtropics (Heinze et al. Citation2006; Schrader et al. Citation2014; Schmidt et al. Citation2016). This ant is an ideal model for studying a wide range of questions of evolutionary biology in eusocial insects, e.g. inbreeding, aging, phenotypic plasticity, and queen competition (Oettler and Schrempf Citation2016). Here, we assembled its complete mitochondrial genome (GenBank: KX951753) from high-throughput sequencing data.
In all, 13.8 M raw reads of 100 bp were retrieved from a previously published study (SRR1564444) (Schrader et al. Citation2014). These data were generated from males of a laboratory-reared colony originally collected from CEPLAC, Ilhéus, Brazil (14°47′36″S, 39°3′9″W). The voucher specimen was stored in the Institute of Zoology, University of Regensburg, Germany. After quality-trimming with Trimmomatic v0.36 (Bolger et al. Citation2014), they were used for mitogenome assembly with the Assembly by Reduced Complexity (ARC) pipeline (https://github.com/ibest/ARC) (Hunter et al. Citation2015), with that of Vollenhovia emeryi (KU550061) (Liu et al. Citation2016) as the reference. Mitogenome annotation was conducted with the MITOS Web Server (Bernt et al. Citation2013), and was delicately adjusted by comparing with those of other ant species (Gotzek et al. Citation2010; Hasegawa et al. Citation2011; Babbucci et al. Citation2014; Duan et al. Citation2016; Kim et al. Citation2016; Liu et al. Citation2016).
The mitochondrial genome of C. obscurior is 15,213 bp long with a highly asymmetric nucleotide composition (38.5% A, 14.3% C, 5.7% G, and 41.5% T; ’light strand‘). It harbors the typical set of 37 animal mitochondrial genes (13 protein-coding genes/PCGs, 22 tRNAs, and two rRNAs) and one control region. The majority of these genes are located on the heavy strand except for four PCGs (nad1, nad4, nad4l, and nad5), eight tRNAs (trnC, trnF, trnH, trnL1, trnP, trnQ, trnV, and trnY), and the two rRNAs (rrnL and rrnS).
The PCGs are initiated with the typical ATA (nad1), ATT (atp8, cox2, nad2, nad3, nad4l, and nad5) or ATG (atp6, cob, cox1, cox3, nad4, and nad6) codons. Three types of stop codons are employed, including TAG (nad4 & nad4l), the incomplete stop codon T(aa) (cox1 and nad1) and TAA (the nine other PCGs). The tRNAs range in size from 59 (trnS1) to 70 bp (trnA) with a total length of 1443 bp. The two adjacent rRNAs are 597 bp (rrnS) and 1367 bp (rrnL) in length, respectively. The control region is 576 bp long with a remarkably high A + T content (90.8%), and is located between rrnS and trnV. This region harbors a 34-bp-long tandem repeat (TA)17 (13,280–13,313). There are twenty intergenic spacer and eight intergenic overlapping regions across the genome.
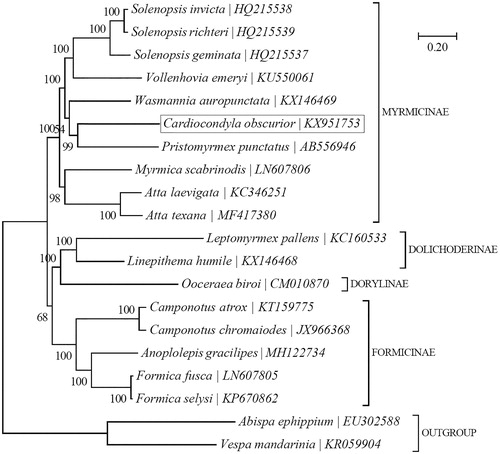
To ascertain its phylogenetic position, a Bayesian tree was reconstructed using the concatenated sequences of 12 well-aligned PCGs (all but atp8) for 18 formicid species with MrBayes v3.1.1 (Ronquist and Huelsenbeck Citation2003) as implemented in TOPALi v2.5 (Milne et al. Citation2009) (). Phylogenetic analysis suggests that C. obscurior is more closely related to Pristomyrmex punctatus than to eight other myrmicine ants.
Figure 1. Phylogenetic relationships among 18 formicid species based on the Bayesian analysis of the concatenated sequences of 12 mitochondrial protein-coding genes (all but atp8; total size: 10,110 bp). The support values are indicated next to the branches. The tree was rooted with two vespid wasps, i.e. Abispa ephippium (EU302588) (Cameron et al. Citation2008) and Vespa mandarinia (KR059904) (Chen et al. Citation2016).
Acknowledgements
The authors thank Dr. Lukas Schrader and his colleagues for generating the genomic data used in this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Babbucci M, Basso A, Scupola A, Patarnello T, Negrisolo E. 2014. Is it an ant or a butterfly? Convergent evolution in the mitochondrial gene order of Hymenoptera and Lepidoptera. Genome Biol Evol. 6:3326–3343.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Cameron SL, Dowton M, Castro LR, Ruberu K, Whiting MF, Austin AD, Diement K, Stevens J. 2008. Mitochondrial genome organization and phylogeny of two vespid wasps. Genome. 51:800–808.
- Chen P-Y, Wei S-J, Liu J-X. 2016. The mitochondrial genome of the Vespa mandarinia Smith (Hymenoptera: Vespidae: Vespinae) and a phylogenetic analysis of the Vespoidea. Mitochondr DNA. 27:4414–4415.
- Duan X-Y, Peng X-Y, Qian Z-Q. 2016. The complete mitochondrial genomes of two globally invasive ants, the Argentine ant Linepithema humile and the little fire ant Wasmannia auropunctata. Conserv Genet Resour. 8:275–277.
- Gotzek D, Clarke J, Shoemaker D. 2010. Mitochondrial genome evolution in fire ants (Hymenoptera: Formicidae). BMC Evol Biol. 10:300.
- Hasegawa E, Kobayashi K, Yagi N, Tsuji K. 2011. Complete mitochondrial genomes of normal and cheater morphs in the parthenogenetic ant Pristomyrmex punctatus (Hymenoptera: Formicidae). Myrmecol News. 15:85–90.
- Heinze J, Cremer S, Eckl N, Schrempf A. 2006. Stealthy invaders: the biology of Cardiocondyla tramp ants. Insect Soc. 53:1–7.
- Hunter SS, Lyon RT, Sarver BAJ, Hardwick K, Forney LJ, Settles ML. 2015. Assembly by Reduced Complexity (ARC): a hybrid approach for targeted assembly of homologous sequences. bioRxiv.
- Kim MJ, Hong EJ, Kim I. 2016. Complete mitochondrial genome of Camponotus atrox (Hymenoptera: Formicidae): a new tRNA arrangement in Hymenoptera. Genome. 59:59–74.
- Liu N, Duan X-Y, Qian Z-Q, Wang X-Y, Li X-L, Ding M-Y. 2016. Characterization of the complete mitochondrial genome of the myrmicine ant Vollenhovia emeryi (Insecta: Hymenoptera: Formicidae). Conservation Genet Resour. 8:211–214.
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 25:126–127.
- Oettler J, Schrempf A. 2016. Fitness and aging in Cardiocondyla obscurior ant queens. Curr Opin Insect Sci. 16:58–63.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Schmidt CV, Schrempf A, Trindl A, Heinze J. 2016. Microsatellite markers for the tramp ant, Cardiocondyla obscurior (Formicidae: Myrmicinae). J Genet. 95:1e–e4.
- Schrader L, Kim JW, Ence D, Zimin A, Klein A, Wyschetzki K, Weichselgartner T, Kemena C, Stökl J, Schultner E, et al. 2014. Transposable element islands facilitate adaptation to novel environments in an invasive species. Nat Commun. 5:5495.
