Abstract
Chicory (Cichorium intybus L.) root is the main raw material used for inulin production, and chicory leaf represents an important forage resource. This is the first reported sequence for a chloroplast genome of the chicory family Compositae. The length of the intact chicory chloroplast genome was 152,975 bp, consisting of one reverse repeat region (IRs, 50,038 bp), one small single-copy region (SSC, 18,561 bp), and one large single-copy region (LSC, 84,376 bp). The chicory chloroplast genome contains 127 functional genes, including 74 protein-coding genes, 29 tRNA genes, and 24 rRNA genes. An analysis of the phylogenetic relationship of chicory to other species revealed that chicory is closely related to the Chrysanthemum and Artemisia groups. Together these results serve to lay a foundation for the ongoing evolutionary and ecological study of Cichorium.
Chicory is a perennial herb of the genus Cichorium in the Compositae family. It is a forage grass of high yield and high quality and is native to the Mediterranean Sea, Central Asia, and North Africa. Chicory has also been used for mixed forage cultivation in grazing lands in New Zealand, Britain, and other countries for more than 300 years (Crush et al. Citation2018). Chicory has a high degree of adaptability and excellent nutritional value, and as such it has become the main form of herbage cultivated in large areas of the United States and Canada (Závada et al. Citation2017). In addition, chicory root is rich in inulin, and as such chicory has become the main crop used for commercial inulin production. Inulin helps slow and reduce the speed and degree of glucose absorption in the body (Ahmed and Rashid Citation2017). However, no Cichorium chloroplast genome sequences have been reported to date. As such, chicory is an excellent material for the study of the evolutionary history of this family of agriculturally important plants. In this study, we provided a theoretical basis for future studies of the genealogical geography of chicory, its phylogenetic evolution, and the relationship between Cichorium and other plants in the Compositae family.
We selected the widely-grown ‘Hera’ variety of chicory as an experimental subject in this study. Fresh leaves were selected, and after snap-freezing, in liquid nitrogen, the RNA was extracted and sequenced by Illumina Hiseq 2500 (Biomarker Technologies, Beijing). SPAdes v3.6.2 was used for sequence splicing (Anton et al. Citation2012). Gap was supplemented by Gapcloser (Luo et al. Citation2015) and GapFiller (Boetzer and Pirovano Citation2012). The obtained sequence information and annotation results were submitted to Genebank with the serial number MK569377. The sequence files in the Genebank format were uploaded to OGDRAW (Lohse et al. Citation2013), and the locations of the IR, LSC, and SSC of chloroplast genome were marked. The double-stranded circular DNA of chicory chloroplast was found to be 152,975 bp in length, containing a reverse repeat region (IR, 50,038 bp), a small single-copy region (SSC, 18,561 bp), and a large single-copy region (LSC, 84,376 bp). The results of chloroplast genome annotation revealed that the chicory chloroplast genome contains 127 functional genes, including 74 protein-coding genes, 29 tRNA genes, and 24 rRNA genes. In addition, it was found that 15 protein-coding genes contained one intron, two genes (ycf3 and clpP) contained two introns, and the GC content of the chloroplast genome was 37.3%.
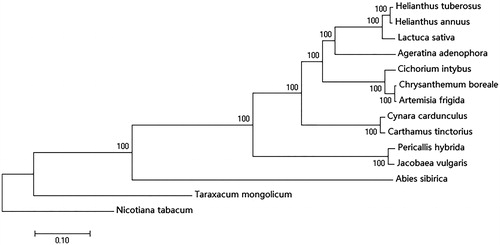
Clustering analysis of 12 chloroplast genomes in the Compositae and Solanaceae family, as well as the chicory genome (), was conducted by Neighbour-joining (Kumar et al. Citation2016). Through this approach, we found that Chrysanthemum boreale, Artemisia frigida, and chicory belong to the same group – a result which will be helpful for the future study of the evolution and ecology of Cichorium. In addition, these data are also an important reference dataset for subsequent studies of Cichorium.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
Figure 1. Neighbour-joining phylogenetic tree of Cichorium intybus based on 14 complete chloroplast genome sequences using Nicotiana tabacum as an out-group. Numbers in the nodes are bootstrap values based on 1000 replicates. Accession numbers are listed as below: Helianthus_tuberosus MG_696658, Helianthus_annuus NC_007977, Lactuca sativa NC_007578, Cynara cardunculus KM_035764, Ageratina_adenophora NC_015621, Chrysanthemum boreale MG_913594.1, Artemisia frigida NC_020607, Cynara cardunculus KM_035764, Carthamus tinctorius KX_822074, Pericallis_hybrida KT_285537, Jacobaea vulgaris NC_015543, Abies sibirica NC_035067, Taraxacum mongolicum NC_031396, and Nicotiana tabacum NC_001879.
Additional information
Funding
References
- Ahmed W, Rashid S. 2017. Functional and therapeutic potential of inulin: a comprehensive review. Crit Rev Food Sci Nutr. 59:1–13.
- Anton B, Sergey N, Dmitry A, Gurevich AA, Mikhail D, Kulikov AS, Lesin VM, Nikolenko SI, Son P, Prjibelski AD. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Boetzer M, Pirovano W. 2012. Toward almost closed genomes with GapFiller. Genome Biol. 13:R56.
- Crush JR, Ouyang L, Cousins GR. 2018. Variation in cadmium concentrations in shoots of chicory (Cichorium intybus L.). New Zealand J Agricult Res. 61:1–9.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2015. Erratum: SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 4:30.
- Závada T, Malik RJ, Kesseli RV. 2017. Population structure in chicory (Cichorium intybus): a successful US weed since the American revolutionary war. Ecol Evol. 7:4209–4219.
