Abstract
The complete mitochondrial genome of Bombus (Mendacibombus) waltoni was sequenced. The circle genome of the bee is 19,349 bp in length. There are 38 sequence elements including 13 protein coding genes, 22 tRNA genes, 2 rRNA genes, and a control region. The order of most elements was consistent with that of other bumblebee species that have been previously described, with the exception of five tRNA genes in two segments. The use of B. waltoni mitogenome as an outgroup significantly improved the robustness of phylogenetic reconstruction for bumblebees.
Bumblebees are a group of insects belonging to the genus Bombus (Hymenoptera: Apidae). There are about 250 known bumblebee species subdivided into 15 subgenera (Williams et al. Citation2008). Most of our current knowledge on bumblebees is heavily biased towards species from subgenus Bombus s. str., especially B. terrestris, while much less is known about other subgenera (Goulson Citation2010). The subgenus Mendacibombus is unique because it is particularly ancient as a phylogenetic group, standing as a sister group to all of the other extant bumblebees (Cameron et al. Citation2007). This root phylogenetic status of Mendacibombus makes it a good reference taxon for phylogenetic reconstruction among bumblebee species. In order to expand the data available for phylogenetic analysis of bumblebees, we reported the mitogenome of B. waltoni, a Mendacibombus species widely distributed on the high elevation regions of China (An et al. Citation2014).
Worker bees were collected from Henan County (N34°37′45″', E101°23′1″ in Qinghai Province, China, and were stored in a −20 °C refrigerator. Genomic DNA was extracted from a single specimen and was sequenced by HiSeq2000 (Illumina), with a read length of 150 bp. The NOVOPlasty v2.7.2 program was used to assemble the mitogenome based on these reads. The assembled sequence was annotated using the web server MITOS (Bernt et al. Citation2013). In order to test the value of our data for phylogenetic analysis of bumblebees, we downloaded the protein-coding genes (PCGs) of all other bumblebee mitogenomes available in GenBank and reconstructed a phylogenetic tree with B. waltoni as an outgroup. The program jModelTest v2.1.6 (Darriba et al. Citation2012) was used to select the best-fitting substitution model. The program MrBayes v3.2.6 (Huelsenbeck and Ronquist Citation2001) was used to reconstruct trees with 1,000,000 generations, which are sufficient to meet the 0.01 criterion of the standard deviation of split frequencies.
A circularized DNA assembly 19,349 bp in length was obtained and deposited in the GenBank with accession number of MK252702. Given the circularity of the sequence and its high (N = 389) average organelle coverage, we posit that this is a complete mitogenome. Similar to other bumblebee mitogenomes, there were 38 sequence elements in this mitogenome: 13 PCGs, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and an AT rich control region (D-loop). All PCGs were distributed similarly as in other Bombus mitogenomes: four were located on the light strand, while others were on the heavy strand (Zhao et al. Citation2017; Takahashi et al. Citation2018).
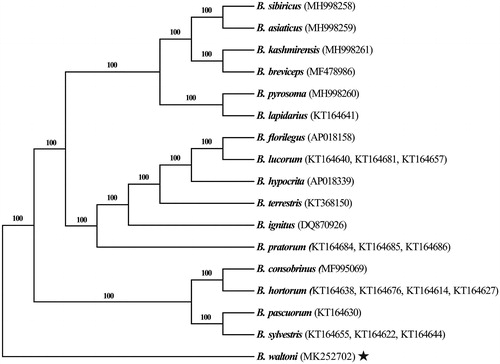
The best-fit nucleotide substitution model GTR + I + G was used for tree reconstruction with B. waltoni as outgroup. The generated Bayesian tree showed accurate interspecific phylogenetic relationships with 100% bootstrap values at all nodes (). As expected, all species assembled into a monophyletic clade within each subgenus. It should be noted that there were 100% bootstrapping rates at all nodes, better than the previous study which used honey bees (Apis spp.) as outgroup species (Takahashi et al. Citation2018). Our results emphasize the value of B. waltoni mitogenomes to phylogenetic analyses of bumblebees.
Figure 1. Phylogenetic analyses of bumblebee mitochondrial genomes with Bombus waltoni as an outgroup. The star indicates the focal mitochondrial genome in this study. Numbers beside each node represent percentages of Bayesian bootstrap values. Species names are followed by the GenBank accession numbers of their mitochondrial genomes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- An JD, Huang JX, Shao YQ, Zhang SW, Wang B, Liu XY, Wu J, Williams PH. 2014. The bumblebees of North China (Apidae: Bombus Latreille). Zootaxa. 3830:1–89.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Cameron SA, Hines HM, Williams PH. 2007. A comprehensive phylogeny of the bumble bees (Bombus). Biol J Linn Soc. 91:161–188.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Goulson D. 2010. Bumblebees: behaviour, ecology, and conservation. Oxford: Oxford University Press.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Takahashi J, Sasaki T, Nishimoto M, Okuyama H, Nomura T. 2018. Characterization of the complete sequence analysis of mitochondrial DNA of Japanese rare bumblebee species Bombus cryptarum florilegus. Conservation Genet Resour. 10:387–391.
- Williams PH, Cameron SA, Hines HM, Cederberg B, Rasmont P. 2008. A simplified subgeneric classification of the bumblebees (genus Bombus). Apidologie. 39:46–74.
- Zhao X, Huang J, Sun C, An J. 2017. Complete mitochondrial genome of Bombus consobrinus (Hymenoptera: Apidae). Mitochondrial DNA Part B. 2:770–772.
