Abstract
IIncarvillea arguta belongs to the family Bignoniaceae, is an economically and medicinally important plant. The complete chloroplast genome of I. arguta is 159,135 bp in size, including a large single copy (LSC) region of 87.192 bp, a small single copy (SSC) region of 19,023 bp and a pair of inverted repeats (IRs) of 26,460 bp. The genome contains 108 genes, including 72 protein-coding genes, 32 tRNA genes, and 4 rRNA genes. The GC content in chloroplast genome, LSC region, SSC region, and IR region were 39.5%, 37.9%, 34.7%, and 43.7%, respectively. The evolutionary analysis suggested that I. arguta is related to Tecomaria capensis.
Incarvillea arguta (Royle) Royle is an economically and medicinally important herb plant, which is widely distributed in the dry-hot valley regions in the Qinghai-Tibetan Plateau and Himalayas. Previous studies had mainly focused on the chemical constituents (Zhou et al. Citation2007), sesquiterpenoids configuration, and biological evaluation (Luo et al. Citation2004; Yan et al. Citation2012) of this species. In this study, we sequenced and assembled the complete chloroplast genome (cpDNA) of I. arguta, which has been deposited into the GenBank with the accession number MG763885.
Fresh leave materials of I. arguta were collected from Xiaojin county, Sichuan, China (N31.01°, E102.04°), and the plant materials and a voucher specimen (No. IAGTX2015067) were stored in Northwest University. Total genomic DNA was isolated using the improved CTAB method (Doyle and Doyle Citation1987), and sequenced with Illumina Hiseq 2500 (San Diego, CA, USA) platform. All raw reads were trimmed using the program NGSQCToolkit_v2.3.3 (Patel and Jain Citation2012). After dislodged the low-quality reads, the clean reads were assembled by MIRA v4.0.2 (Chevreux et al. Citation2004) and MITObim v1.8 (Hahn et al. Citation2013) using the chloroplast genome of Tanaecium tetragonolobum (NC_027955) as a reference.
The chloroplast complete genome of I. arguta is 159,135 bp in size, including a large single copy (LSC) region of 87.192 bp, a small single copy (SSC) region of 19,023 bp and a pair of inverted repeats (IRs) of 26,460 bp. The GC content in chloroplast genome, LSC region, SSC region and IR region were 39.5%, 37.9%, 34.7%, and 43.7%, respectively. The genome contains 108 genes, including 72 protein-coding genes, 32 tRNA genes, and 4 rRNA genes.
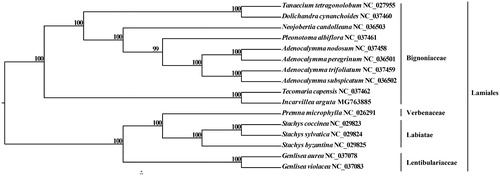
A total of 16 species were used to construct the phylogenetic tree among the most of Lamiales species with Verbenaceae, Lentibulariaceae, and Labiatae species as outgroups. Modeltest v3.7 (Posada and Crandall Citation1998) was used to determine the best-fitting model for the chloroplast genomes. Maximum likelihood analysis was performed using RAxML (Stamatakis Citation2014) with 1000 bootstrap replicates. The results showed that I. arguta is closely related to Tecomaria capensis (). The complete chloroplast genome of I. arguta can be subsequently used for the phylogenetic reconstruction and comparative genomics analysis of Bignoniaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Luo Y, Yi J, Li B, Zhang G. 2004. Novel ceramides and a new glucoceramide from the roots of Incarvillea arguta. Lipids. 39:907–913.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619
- Posada D, Crandall KA. 1998. Modeltest: Testing the model of DNA substitution. Bioinformatics. 14:817–818.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Yan YM, Wu GS, Dong XP, Shen L, Li Y, Su J, Luo HR, Zhu HJ, Cheng YX. 2012. Sesquiterpenoids from Incarvillea arguta: absolute configuration and biological evaluation. J Nat Prod. 75:1025.
- Zhou DY, Yang XS, Yang B, Zhu HY, Hao XJ. 2007. Chemical Constituents of Incarvillea arguta. Nat Prod Res Dev. 19:807.