Abstract
The complete mitochondrial genome of a lower termite, Reticulitermes leptomandibularis is a typical circular DNA molecule of 15,920 bp in length. The genome contains 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and a non-coding region (D-loop).The overall sequence of R. leptomandibularis is A/T biased with a percentage of 65.36%. The phylogenetic tree constructed using complete mitochondrial genomes of Reticulitermes genus revealed that R. leptomandibularis was closest to Reticulitermes aculabialis and formed a sister group to Reticulitermes speratus. This whole mt genome provides molecular resource for evolution analysis within termites especially the genus Reticulitermes.
The genus Reticulitermes contains some significant pest species in the world (Cameron and Whiting Citation2007). Reticulitermes leptomandibularis is an economically important species in China with obvious morphological characteristics of Reticulitermes. Although it was first reported in 1965 (Hsia and Fan Citation1965), there is no information about the complete mitochondrial genome of this species. This study provides the complete mitogenome sequence of R. leptomandibularis.
Specimens of R. leptomandibularis were collected from Hangzhou, Zhejiang, China and kept in the insect laboratory of Zhejiang A&F University (accession number ZJ0001-LA-1). The complete mitogenome sequence of R. leptomandibularis was 15,920 bp in length with Genbank accession number MK541931. The mitogenome contains 13 PCGs (nad1-6, nad4L, atp6, atp8, cox1-3, cytb), 22 tRNA genes, 2 rRNA genes (rrnL and rrnS), and a non-coding control region (D-loop). The gene content and orientation were identical to those previously reported from other Reticulitermes species (Cameron and Whiting Citation2007; Chen et al. Citation2016; Lee et al. Citation2017). The H-strand contained 9 PCGs and 14 tRNAs while the remaining genes including 4 PCGs, 8 tRNAs, and 2 rRNAs were located on L-strand.
The overall nucleotide composition of R. leptomandibularis mtDNA was significantly A/T biased (65.36%). The mitogenome of R. leptomandibularis included intergenic spacers and overlapping regions. The intergenic spacer sequences were 112 bp in total and were located in 18 regions, which varied from 1 to 17 bp in length. There were 29 bp overlapping sequences varied from 1 to 8 bp in 6 areas.
The total 13 PCGs was 11,168 bp in length. PCGs were initiated with an ATN start codon. All PCGs contained standard stop codon TAA except cox2 and nad5 were terminated with a single T and nad1 was terminated with TAG. All 22 tRNAs displayed a typical clover-leaf structure, except for tRNASer(AGN). As typically observed in insect mitogenomes, two rRNA genes (rrnL and rrnS) were found on the L-strand with 1303 bp and 727 bp in length, respectively. The control region (D-loop) was 1124 bp in length located between rrnS and trnL gene. The D-loop is also A/T biased with a percentage of 67.79%, which was higher than that of the overall mtDNA.
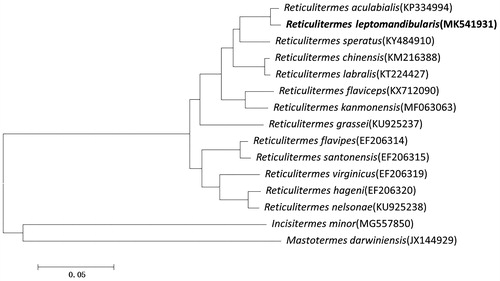
A phylogenetic analysis was performed based on 13 PCGs nucleotide sequences from 13 closely related Reticulitermes species including R.leptomandibularis using a maximum likelihood method (Guindon et al. Citation2010). Incisitermes minor and Mastotermes darwiniensis were chosen as outgroups for phylogenetic tree. The phylogenetic analysis showed there were two major clades formed, one is a clustering of I. minor and M. dareiniensis and the other was Reticulitermes species (). The phylogenetic tree revealed that R. leptomandibularis was closest to R. aculabialis and constituted a sister group to R. speratus. The data provided a useful resource especially for classification in genus Reticulitermes.
Nucleotide sequence accession number
The complete genome sequence of R. leptomandibularis has been assigned GenBank accession number (MK541931).
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Cameron SL, Whiting MF. 2007. Mitochondrial genomic comparisons of the subterranean termites from the Genus Reticulitermes (Insecta: Isoptera: Rhinotermitidae). Genome. 50:188–202.
- Chen Q, Wang K, Tan Y, Xing LX. 2016. The complete mitochondrial genome of the subterranean termite, Reticulitermes chinensis Snyder (Isoptera: Rhinotermitidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1428–1429.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Hsia KL, Fan ST. 1965. Notes on the genus Reticulitermes Holmgren of China(Isoptera, Rhinotermitidae). Acta Entomologica Sinica. 14:360–382.
- Lee W, Han T, Lee J-H, Hong K-J, Park J. 2017. The complete mitochondrial genome of the subterranean termite, Reticulitermes speratus kyushuensis Morimoto, 1968 (Isoptera: Rhinotermitidae). Mitochondrial DNA B. 2:178–179.