Abstract
Otters are flagship species for pristine habitats and their southernmost distribution in Africa includes two species; Aonyx capensis and Hydrictis maculicollis. Here, we present novel full mitochondrial genomes of these otter species. The comparable mitogenomes consist of 36 genes including 13 protein-coding genes, 2 ribosomal RNAs, and 22 tRNAs including a hypervariable region. Only 19 out of the 36 genes showed some level of variation between species with the smallest being trnV (68 bp difference) and the biggest being nad5 (1830 bp difference). Such variations may provide guidance in selecting gene regions during marker development for phylogenetic assessments.
Otters (Lutrinae) belong to the largest family of carnivores: Mustelidae (Koepfli et al. Citation2008). In South Africa, there are two species of otter; spotted-necked (Hydrictis maculicollis) and African clawless (Aonyx capensis). The International Union for Conservation of Nature (IUCN) list these otters as Near Threatened (Jacques et al. Citation2015; Reed-Smith et al. Citation2015). In this study, we for the first time sequenced, annotated, and compared mitogenomes of two South African otters.
Specimens were obtained from opportunistic sampling for A. capensis (approximate coordinates: 34°02′06.5′′S 23°01′27.5′′E) and H. maculicollis (approximate coordinates: 28°43′43.3′′S 20°59′09.5′′E) and are stored at the National Zoological Garden (NZG), South African National Biodiversity Institute (SANBI ) Biobank at −20 °C. DNA was extracted using the ZymoResearch Quick-DNA Tissue kit (Zymo Research Corp., Irvine, CA). Sequencing was performed on an Illumina HiSeq 2500 (Illumina Incorporated, San Diego, CA). Sequencing produced 2738699 and 2840359 reads for A. capensis and H. maculicollis, respectively, which were assessed for quality using FastQC version 0.11 (BaseSpace Labs App., Illumina Incorporated, San Diego, CA). Editing and trimming of data were done using Trimmomatic version 0.36 (USADEL LAB, Aachen, Germany). Single end read assembly of both genomes was done De Novo, using CLC Genomics Workbench version 6 (CLC Bio, Aarhus, Denmark). The number of reads successfully mapped were 3545 (38 × coverage) and 82244 (850 × coverage) for A. capensis and H. maculicollis, respectively. Annotation was performed using MITOS webserver version 806 (University of Leipzig, Leipzig, Germany) and the BLAST Ring Image Generator (Alikhan et al. Citation2011) was used to perform circular alignments.
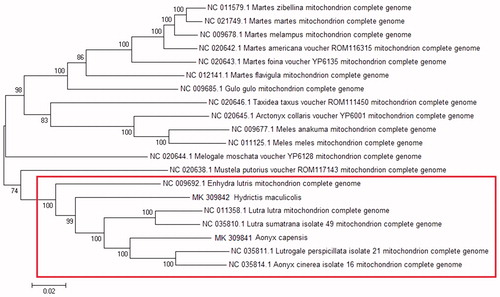
The complete mitochondrial genomes of A. capensis (16,188 bp) and H. maculicollis (16,308 bp) are comparable in length to other otter species. The arrangement of genes is also similar to otters and other mustelid species, 36 genes including 13 protein-coding genes, 2 ribosomal RNAs, and 22 for tRNAs including a hypervariable region. Of these, 27 are codons on the sense (+) strand with the remaining nine on the antisense (−) strand for both species. The 13 protein-coding genes share similar start/stop codons between species with variations only observed at gene nad2 (start codon), cox2 (stop), and nad3 (start) where the codons are ATT, TAA, and ATT for H. macullicolis and ATC, TAG, and ATA for A. capensis, respectively. Only 19 out of the 36 genes showed some level of variation between the two species with the smallest being trnV with a 68 bp difference and the biggest being nad5 with an 1830 bp difference. Individual GC content calculations for both A. capensis and H. maculicollis gave 42.7 and 41.3%, respectively which are comparable to a variety of mammals, such as brown bears (41.3%), Eurasian otter (42.1%), and sea otter (41.1%) (Ki et al. Citation2010). To explore phylogenetic relationships, a maximum likelihood (ML) tree was constructed using available mitogenomes from mustelid species. This phylogenetic reconstruction was performed using the Molecular Evolutionary Genetics Analysis (MEGA) phylogenetic software version 7.0.9 (Pennsylvania State University, State College, PA) (Kumar et al. Citation2016). The topological structure strongly supports the published taxonomy (Koepfli et al. Citation2008) of the Lutrinae family of mustelids with a bootstrap probability of 100% in almost all clades (). The two mitogenomes will provide invaluable genetic resources for further studies.
Acknowledgements
We would like to acknowledge Roger Bills at the South African Institute for Aquatic Biodiversity (SAIAB) for the H. maculicollis specimen and Coral Birss of CapeNature for the A. capensis specimen.
Disclosure statement
The authors report no conflicts of interest and are solely responsible for the content and writing of this manuscript.
Additional information
Funding
References
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 12:402.
- Jacques H, Reed-Smith J, Somers MJ. (2015). Aonyx capensis. The IUCN Red List of Threatened Species Retrieved from www.iucnredlist.org. Accessed 1 December 2018.
- Ki JS, Hwang DS, Park TJ, Han SH, Lee JS. 2010. A comparative analysis of the complete mitochondrial genome of the Eurasian otter Lutra lutra (Carnivora; Mustelidae). Mol Biol Rep. 37:1943–1955.
- Koepfli KP, Deere KA, Slater GJ, Begg C, Begg K, Grassman L, Lucherini M, Veron G, Wayne RK. 2008. Multigene phylogeny of the Mustelidae: resolving relationships, tempo and biogeographic history of a mammalian adaptive radiation. BMC Biol. 6:10.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Reed-Smith J, Jacques H, Somers MJ. (2015). Hydrictis maculicollis. The IUCN Red List of Threatened Species 2015. Retrieved from www.iucnredlist.org. Accessed 1 December 2018.