Abstract
Aristobia reticulator (Fabricius, 1781) is an important coleopteran pest of guava and litchi in India and China. The draft mitochondrial genome (mitogenome) of A. reticulator is ca. 15,838 bp long and consists of 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, 22 transfer RNA (tRNA) genes, and a control region (MK423971). The nucleotide composition of the mitogenome was significantly A + T-biased (A: 39.3%, T: 39.2%, G: 8.7%, C: 12.8%). All 22 tRNA genes show typical secondary cloverleaf structures. The mitogenome gene content and gene order was as described for a majority of coleopteran species. Phylogenetic analysis revealed that A. reticulator was closely related to Anoplophora chinensis (Forster), Anoplophora glabripennis (Motschulsky), and Anoplophora lurida (Pascoe, 1857) of the coleopteran tribe Lamiini (Cerambycidae, Lamiinae).
The coleopteran insect family “Cerambycidae” includes many species that are of agricultural and horticultural importance. Globally, over 50 species of cerambycids are well-known pests of agricultural, horticultural, and forest ecosystems; and a majority of these are borers of stem, shoot, trunks and roots of plants. A total of 163 species of longicorn beetles are known from Indian’s northeastern region which forms part of the Indo-Myanmar mega-diversity hotspot (Agarwala and Bhattacharjee Citation2015). Considered as an important pest of guava and litchi, A. reticulator is known as guava trunk borer (Shylesha et al. Citation2000), litchi stem borer or litchi longhorn beetle (Xu et al. Citation1995; Peng et al. Citation2003), and has a wide geographic distributional range from Nepal to Vietnam (Kumawat et al. Citation2015). Despite its economic importance, there has been no molecular characterization for A. reticulator including a complete lack of the mitochondrial DNA cytochrome oxidase subunit I (mtCOI) gene that is widely used in DNA barcoding to assist with confidence in species identification.
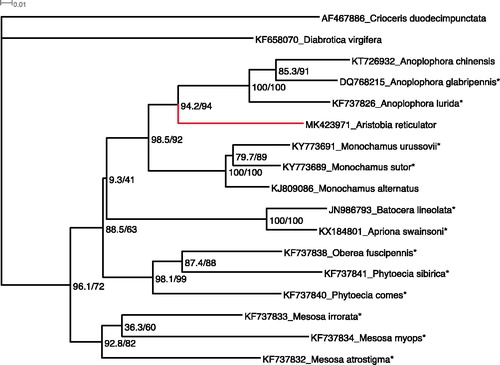
In this report, we sequenced and characterized the mitogenome of an adult A. reticulator collected from Guava plants grown at Umiam (25°.677′N, 91°.927′E), Meghalaya state of India in August 2014. Total genomic DNA was extracted from muscle tissues of the adult individual using standardized isolation kit. One specimen each of Male and Female of Aristobia reticulator have been deposited as voucher specimens in the Insect Museum of Division of Crop Protection, ICAR Research Complex for NEH Region, Umiam (Meghalaya), India. The paired-end sequencing library was prepared using Illumina TruSeq Nano DNA HT Library Preparation Kit. The complete genome of A. reticulator was sequenced using NextSeq platform (Xcelris Labs Ltd., Ahmedabad, India). We assembled the A. reticulator mitogenome using Geneious v11.1.5 (Biomatters Ltd., Auckland, New Zealand) and annotation using Mitos (Bernt et al. Citation2013), with fine-tuning carried out in Geneious. Although coleopteran mitogenome gene rearrangements are known (Andujar et al. Citation2016), the gene order and orientation of the A. reticulator mitogenome was identical when compared with the majority of published coleopteran mitogenomes (e.g. Behere et al. Citation2016) especially with that of the Cerambycidae (Li et al. Citation2016; Liu et al. Citation2018). Maximum Likelihood phylogenetic analysis using the mtCOI gene of A. reticulator and selected Cerambycidae species was carried out using IQ-TREE (Nguyen et al. Citation2015; Trifinopoulos et al. Citation2016) based on the parameters reported in Kunz et al. (Citation2019) that showed A. reticulator as basal to Anoplophora species with strong SH-aLRT and ultrafast bootstrap support values ().
Figure 1. Maximum likelihood inferred phylogenetic relationship of Aristobia reticulator (Fabricius, 1781) (red colour branch) against selected Cerambycidae beetles in the subfamily Lamiinae based on 1,543 bp of mitochondrial DNA COI gene, with Diabrotica virgifera virgifera and Crioceris duodecimpunctata (Linnaeus) (Chrysomelidae) as outgroups. Phylogeny was inferred using IQ-TREE with automatic evolutionary model selection and 1,000 bootstrap replications to assess for node support. Node values are SH-aLRT support (%) / ultrafast bootstrap support (%), where values of >80% / ≥95% can be considered as strong support for clades, respectively. Unpublished mtDNA COI gene sequences obtained from GenBank are indicated by ‘*’.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Agarwala BK, Bhattacharjee PP. 2015. Redescription of Aristobia reticulator (F., 1781) (Coleoptera: Lamiinae), with a taxonomic note and record of a new food plant for adults in Northeastern India. Coleopt Bull. 69:205–212.
- Andujar C, Arribas P, Linard B, Kundrata R, Bocak L, Vogler AP. 2016. The mitochondrial genome of Iberobaenia (Coleoptera: Iberobaeniidae): first rearrangement of protein-coding genes in the beetles. Mitochondrial DNA Part A. 28:156–158.
- Behere GT, Firake DM, Tay WT, Azad Thakur NS, Ngachan SV. 2016. Complete mitochondrial genome sequence of a phytophagous ladybird beetle, Henosepilachna pusillanima (Mulsant) (Coleoptera: Coccinellidae). Mitochondrial DNA Part A. 27:291–292.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler P. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Kumawat MM, Mamocha Singh K, Ramamurthy VV. 2015. A checklist of the long-horned beetles (Coleoptera: Cerambycidae) of Arunachal Pradesh, Northeastern India with several new reports. J Threat Taxa. 7:7879–7901.
- Kunz D, Tay WT, Court LN, Elfekih S, Gordon KHJ, Evans GA, De Barro PJ. 2019. Draft mitochondrial DNA genome of a 1920 Barbados cryptic Bemisia tabaci ‘New World’ species (Hemiptera: Aleyrodidae). Mitochondrial DNA Part B. 4:1183–1184.
- Li W, Yang X, Qian L, An Y, Fang J. 2016. The complete mitochondrial genome of the citrus long-horned beetle, Anoplophora chinensis (Coleoptera: Cerambycidae). Mitochondrial DNA Part A. 27:4665–4667.
- Liu YQ, Chen DB, Liu HH, Hu HL, Bian HX, Zhang RS, Yang RS, Jiang XF, Shi SL. 2018. The complete mitochondrial genome of the longhorn beetle Dorysthenes paradoxus (Coleoptera: Cerambycidae: Prionini) and the implication for the phylogenetic relationships of the Cerambycidae species. J Insect Sci. 18:21.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Peng H, Liu R, Lu J. 2003. The variety of resources and cultivation of lychee in China. Proc Florida State Hort Soc. 116:1–3.
- Shylesha AN, Azad Thakur NS, Ramchandra A. 2000. Incidence of litchi borer, Aristobia testudo Voet (Coleoptera: Lamiidae) on guava in Meghalaya. Pest Manag Hort Ecos. 6:156–157.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucl Acids Res. 44:W232–W235.
- Xu J, Han R, Liu X, Cao L, Yang P. 1995. Application of codling moth nematode Steinernema carpocapsae against the larvae of litchi longhorn beetle Aristobia testudo. Acta Phytophylacica Sinica. 22:12–16. (in Chinese).
