Abstract
The mitochondrial DNA (mtDNA) has been widely used as a suitable tool for phylogenetic studies among species, including molecular evolution, phylogenetics, population genetics, and comparative and evolutionary genomics. We described the complete mitogenome of Dysomma anguillare in this study. The mitogenome is 16,689 bp in length and contains 13 protein-coding genes (PCGs), 2 rRNA genes (12S rRNA and 16S rRNA), 22 tRNA genes, and a putative control region (CR) and one origin of replication (OL) on the light-strand. Moreover, a phylogenetic tree based on the neighbor-joining method was constructed to explore the phylogenetic relationship of D. anguillare, which strongly supported it had the closest relationship with the Simenchelys parasitica. These results would provide comprehensive phylogenetic relationship within Gobionellinae and additional information for management for this species.
Keywords:
The Dysomma anguillare (shortbelly eel or arrowtooth eel) is an eel in the family Synaphobranchidae (cutthroat eels), which is a marine, tropical eel and known from the western Atlantic Ocean and Indo-Western Pacific, including the United States, Venezuela, South Africa, Zanzibar, and Japan (Ho et al. Citation2015). This study determined the complete mitochondrial genome of D. anguillare to facilitate future studies on population genetic structure and phylogenetic relationships.
The specimen was collected from Zhoushan city, China (29°59′3729° 122°12′28″82 and stored in the laboratory of Zhejiang Ocean University with accession number QGM-01. The complete mitogenome of D. anguillare is 16,689 bp long (GeneBank Accession No. MK189460) and contains 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, 22 transfer RNA genes, and 2 main non-coding regions, this feature was similar to the typical mitogenome of other vertebrates (Miya et al. Citation2001; Zhu, Gong, Jiang, et al. Citation2018; Zhu, Gong, Lü, et al. Citation2018). The overall base composition is 33.2% A, 28.2% C, 23.0% T, and 15.6% G. Twelve PCGs, fourteen tRNA genes, and two rRNA genes were located on the heavy strand, while one PCG (ND6) and eight tRNA genes (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro) on the light strand. The 13 PCGs genes encode 3813 amino acids in total. All the PCGs use the initiation codon ATG except COI uses GTG, which is quite common in vertebrate mtDNA (Balakirev et al. Citation2016; Du et al. Citation2018; Zhu, Lü, et al. Citation2018). Most of them have TAA or TAG as the stop codon, except two PCGs (COII and ND4) use AGA, and COI ended with a single T, these incomplete termination codons were presumably completed as TAA by post-transcriptional polyadenylation (Ojala et al. Citation1981; Zeng et al. Citation2012). The two ribosomal RNA genes, 12s rRNA (956 bp), and 16s rRNA (1709 bp) are located between tRNA-Phe and tRNA-Leu (UUA) and are separated by the tRNA-Val gene on the heavy strand. All 22 tRNA genes of D. anguillare can fold into a typical cloverleaf structure except tRNA-Ser (AGC), which lacks a dihydrouridine arm. By comparing the sequences of other teleosts, the OL is topically located in a cluster of five tRNA genes (Trp, Ala, Asn, Cys, and Tyr) as in other vertebrates, which can fold into a stem-loop secondary structure with a stem formed by 11 paired nucleotides and a loop of 10 nucleotides. The CR, which is determined to be 960 bp, is located between tRNA-Pro and tRNA-Phe, and the central and conserved sequence blocks (CSB-1, CSB-2, and CSB-3) were identified (Cheng et al. Citation2012).
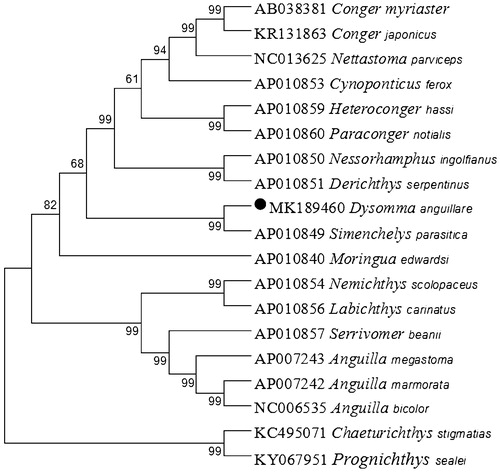
We performed bootstrap analyses (1000 replicates) to evaluate relative levels of support for various nodes in the phylogeny (). The NJ tree indicated that D. anguillare has the closest relationship with Simenchelys parasitica. The results of this study could serve as a basis for future studies on the phylogenetic relationships of this species.
Figure 1. Neighbor Joining (NJ) tree of 17 Anguilliformes species based on 12 PCGs encoded by the heavy strand. The bootstrap values are based on 1000 resamplings. The number at each node is the bootstrap probability. The number before the species name is the GenBank accession number. The genome sequence in this study is labeled with a black spot.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Balakirev ES, Saveliev PA, Ayala FJ. 2016. Complete mitochondrial genome of the Amur sculpin Cottus szanaga (Cottoidei: Cottidae). Mitochondr DNA B. 1:737–738.
- Cheng Y, Xu T, Jin X, Tang D, Wei T, Sun Y, Meng F, Shi G, Wang R. 2012. Universal primers for amplification of the complete mitochondrial control region in marine fish species. Mol Biol. 46:727–730.
- Du X, Gong L, Chen W, Liu L, Lü Z. 2018. The complete mitochondrial genome of Epinephelus chlorostigma (Serranidae; Epinephelus) with phylogenetic consideration of Epinephelus. Mitochondr DNA B. 3:209–210.
- Ho HC, Smith DG, Tighe KA. 2015. Review of the arrowtooth eel genera Dysomma and Dysommina in Taiwan, with the description of a new species (Anguilliformes: Synaphobranchidae: Ilyophinae). Zootaxa. 4060:86.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18:1993–2009.
- Ojala D, Montoya J, Attardi G. 1981. TRNA punctuation model of RNA processing in human mitochondrial. Nature. 290:470–474.
- Zeng Z, Liu ZZ, Pan LD, Tang SJ, Wang CT, Tang WQ, Yang JQ. 2012. Complete mitochondrial genome of the Endangered roughskin sculpin Trachidermus fasciatus (Scorpaeniformes, Cottidae)). Mitochondrial DNA. 23:435–437.
- Zhu K, Gong L, Jiang L, Liu L, Lü Z, Liu BJ. 2018. Phylogenetic analysis of the complete mitochondrial genome of Anguilla japonica (Anguilliformes, Anguillidae). Mitochondr DNA B. 3:536–537.
- Zhu K, Gong L, Lü Z, Liu L, Jiang L, Liu B. 2018. The complete mitochondrial genome of Chaetodon octofasciatus (Perciformes: Chaetodontidae) and phylogenetic studies of Percoidea. Mitochondr DNA B. 3:531–532.
- Zhu K, Lü Z, Liu L, Gong L, Liu B. 2018. The complete mitochondrial genome of Trachidermus fasciatus (Scorpaeniformes: Cottidae) and phylogenetic studies of Cottidae. Mitochondr DNA B. 3:301–302.
